













| General Information: |
|
| Name(s) found: |
SPAC1D4.10
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 809 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
tRNA 3'-trailer cleavage
[ISS |
| Molecular Function: |
zinc ion binding
[IEA]
3'-tRNA processing endoribonuclease activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKTVNFRAT KNFYLQFVSV SSRDTSCIPC IHLFFDSKRY VFGSVGEGCQ RAILSQQLRL 60
61 SKIKDVFLMQ GSSISSPDTY DSSSSSSTTS VSDMLQLDDR DKVIVSERNS MCSTVNYPTW 120
121 WDSCGGFPGF LLSLNDISEP GETGEASPFV LHGPSEVHQF LSSMRHFTYH TNVNLTVQGY 180
181 TSAEAPVFVD ENICVTPVVV SLVKNSFKKR KHENINRGTN ARPLKEDRAN TSPHWYSHVS 240
241 NDTSFVVENA MYNTPAPLEP DKPELFISYI VQSHPTPGKF DAAKAKSLGI TKGLDCGRLA 300
301 RGEPVTLENG KTVYPKEVIG PSIPGSSFFI IHCPNELVID LVIENHKWTN APKPVCVIHS 360
361 VTPEVYKNPR YQSWISSFPS EVSHLIASTE VNEVINYPRS AVAIATLNLL DSKVFPLGFN 420
421 CYEVKNVQKN NRIAFAKPKL RFAFGKKTGI DDSEVGVSIE ELKDKILKEK PDYKSFVEEA 480
481 QKYVSDKPKA PSFAGSDIQI CTLGTGSAMP SLYRNVSSTY VRIPVDKKCM EDSAISMKNI 540
541 LLDCGEGTLG RLSRQYGDNL KYEIASLRWI YISHMHADHH AGVIGVLKAW TKYSDGRSKL 600
601 FITAPPQFEF WLLEYSRIDY LPLSNIVFIS NSALRTDRKP SALESSRLSS LFKEFDLVSF 660
661 RTVPAIHCPY SYCMEITNSS GWKIAYSGDT RPSEDFANIA KDSTLLIHEA TLEDSMHEIA 720
721 IKKQHSTYSE ALEVAKKAGT KNVILTHFSQ RYPKLPDIDI STEDLHIALA FDGMTLKISD 780
781 ISLFRYFGKP LAYLFNEENL KEESDPLKF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
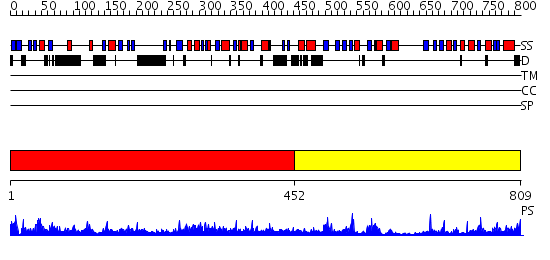
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..451] | 47.69897 | Crystal structure of RNase Z | |
| 2 | View Details | [452..809] | 34.045757 | Crystal Structure of the Coenzyme PQQ Synthesis Protein (PqqB) from Pseudomonas putida, Northeast Structural Genomics Target PpR6 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)