













| General Information: |
|
| Name(s) found: |
ubp2 /
SPAC328.06
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1141 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
protein deubiquitination
[ISS ubiquitin-dependent protein catabolic process [IEA] |
| Molecular Function: |
ubiquitin-specific protease activity
[ISS ubiquitin thiolesterase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATVKNLRIG KSPNRLIQDL DVFDSPSAGW NDPWSPHSSR YHWQLHSNLG ESAVFDENDF 60
61 WCVCQKTRKH LHVKVRRDRG KPLIEEPEEN YGIKDRIPVY YEEEELPEPH VTSPTKSEFA 120
121 TTSTCMKWSS KTTKSEIEVE WRDSYLDANC IKEIIDSRRP SFASLLTKKS SSHQGSSHSS 180
181 QPSLFTTFTS LELFLRNVLV HNDQRAISAA PEGTFERHVG KGRQIQSLMK SLLFEYHHEN 240
241 VNYVPTIADA PLTDEQKLNL YLARNELIVL ANHFRDTKED PAIVANPFPV RLARPALINA 300
301 FGVPNYDSVV PMYTTVFRDN SASLPDDPAF IALGITNDYP DSFVRYFYEE QKKNDEANVR 360
361 VYADALAHIY NLRKSSFLRD LIAADRKNGI VSSDVIQAAY SSLGLEAEVG PDYRYSQEKI 420
421 FEAFHSALLR KPEFARAIRN DLETIGYARK SSEILNYVLS TEQAFYTVNE AYQWLGIKSN 480
481 TEDAMVASVA LVKFEDDSDK AIEAVKWIAE ERNSSILYDF LASQGRPSNK KPKEVPMDED 540
541 LAYNTLGVQD RALSDDVLIN VYGFAVEDHP EQSDTLRAAL KCIGEVRNSR LITHYLEHGN 600
601 LDIPPEVSSL DTPIGLENTG NLCYLNSLIQ YYFIIKPLRN AILDIDENKD LNMIENKEAV 660
661 KKVGGRIVTR IEFLRALQFT YELRKLFIEL ITSKSSSVHP SSVLTYLALI PLTLDQVKSG 720
721 TSSVMDLSSS RELSNLNERS ITIDPRAEEQ AQGLEQEQGQ DEAKSPAEQS SSVNLIDFPM 780
781 ANTNGESQTQ PHYFEVSEEE INSSMDLGRQ QDVLECIDHV LFQLEASLGR ISNSEDRLGS 840
841 DNDLIRRLFS GKLKQTLNDA SQGVRSNYEI FSHLIVDLFE EKQTLYDALD GVFETVNIDM 900
901 GSETAQRSLC ITELPIILQL QIQRVQFDRT TGQPFKSNAF VEFGKELSMD RYVEDTDGKM 960
961 APLLQRYWDL KREIINLQKR QQLLLTTNSN LMSSVDTLSI LSKWAAQQQD SRLPINPKLP1020
1021 DILQEEINNV VAEVDMLKKQ EASLKEERTH LFDNYISHSY DLLAVFVHRG QASFGHYWTY1080
1081 IHDFENNVYR KYNDEYVTVV DESEIFADTT GNNANPYMLT YIRKEYRHII ECVHREHNLL1140
1141 L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
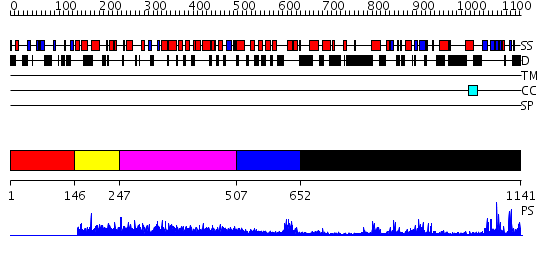
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..145] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [146..246] | 1.38 | No description for 2d5uA was found. | |
| 3 | View Details | [247..506] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [507..651] | 1.49 | No description for 2gfoA was found. | |
| 5 | View Details | [652..1141] | 25.69897 | Crystal structure of HAUSP |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)