













| General Information: |
|
| Name(s) found: |
ntp1 /
SPBC660.07
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 735 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
ascospore formation
[IMP response to cold [IDA cellular response to heat [IDA response to osmotic stress [IDA cellular response to oxidative stress [IDA trehalose metabolic process [TAS trehalose catabolic process [IEA] |
| Molecular Function: |
protein binding
[IPI calcium ion binding [IDA alpha,alpha-trehalase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPSKFSSKYV DTEAISNDDD NPFATAKSYY SKDTDLSTRV SAGRPRTLST SMEASAAPTI 60
61 PELKNLRRRG SLDEHKQPRK FLVDVDKTLN ALLESEDTDR NMQITIEDTG PKVVSLGSAS 120
121 SGGYRLYELR GTYQLSNLLQ ELTLAKDYGR RYILLDERRL NENPVNRLSR LIKGTFWDAL 180
181 TRRIDASVLD VICRDTKDRS GSHVNRIYVP KAEQEMYEYY VRAAKERPYL NLQVEYLPEE 240
241 ITPEWVRDVN DKPGLLALAM EKYQDDEGNT HLRGVPFVVP GGRFNELYGW DSYFESLGLL 300
301 VDDRVDLAKG MVENFIFEIT YYGKILNANR TYYLLRSQPP FLTDMALRVY ERIKNEEGSL 360
361 DFLHRAFSAT IKEYHTVWTA TPRLDPKTGL SRYRPGGLGI PPETEASHFE HLLRPYMEKY 420
421 HMTLEEFTHA YNYQQIHEPA LDEYFVHDRA VRESGHDTTY RLEKVCADLA TVDLNSLLYK 480
481 YETDISHVIL EYFDDKFVLP NGTIETSAIW DRRARARRAA MEKYLWSEAD SMWYDYNTKL 540
541 ETKSTYESAT AFWALWAGVA TPRQAAKFVD VSLPKFEVAG GIVAGTKRSL GKVGLDNPSR 600
601 QWDYPNGWSP QQILAWYGLI RYGYEEETRR LVYRWLYTIT KSFVDFNGIV VEKYDLTRPV 660
661 DPHRVEAEYG NQGVNIKGVA REGFGWVNAS YEVGLTFCNS HMRRALGACT TPDVFFAGIK 720
721 EESLPAFENL SIHKN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
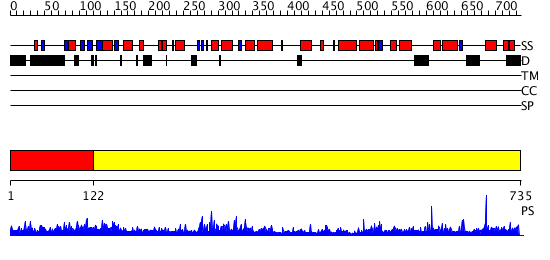
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [122..735] | 103.0 | No description for 2jf4A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)