













| General Information: |
|
| Name(s) found: |
tpp1 /
SPAC19G12.15c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 817 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
cellular response to stress
[IEP trehalose biosynthetic process [IEA] trehalose metabolic process [IMP dephosphorylation [IC |
| Molecular Function: |
protein binding
[IPI trehalose-phosphatase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVYGKIPST SFEHENTFEL SGDLLDPEEL KSLGVSGRII YVLRHLPFKS SINEETREWD 60
61 LSGRRGATTM YSSMNWLANS TYWQTTLVGW TGVIPTVSEK EENKDAVTRL DSQDVKRFEE 120
121 TYSQWNSGER STEYVPVWLP GPEKGSETII NETRSQQSRW LAYAENVIRP LIHYKYWPSS 180
181 EVDENEEQWW RDYVKMNHAF ADKICEIYKP GDFIIVQDYS LFLVPQLIRN KIDDAVIGFY 240
241 HHHPFPSSEI ARCFPRRRAI LRSVLGADFI GFEDYSYARH FISCCSRVLD LEIGHDWVNL 300
301 NGNKVTVRAI TVGIDVPRII RSSGNVSVSE KLEELNKRYE NMKVILGRDR LDELYGVPQK 360
361 LRSFQRFLRT YPEWRKKVVL IQITISSAFK HPKLLSSIKK LVQAINQEFG TDDYTPVHHV 420
421 EEQLEPADYF ALLTRADALF INSIREGVSN LALEYVVCQR DRYGMVLLSE FTATSAMLHD 480
481 VPLINPWDYN ECAEIISNAL STPLERRKMI ERESYKQVTT HTMQSWTSSL IRSLANKLAA 540
541 TKTDQRIPTL TPEHALSVYS KASKRLFMMD YDGTLTPIVR DPNAAVPSKK LLDNLATLAA 600
601 DPKNQVWIIS GRDQQFLRNW MDDIKGLGLS AEHGSFVRKP HSTTWINLAE LLDMSWKKEV 660
661 RRIFQYYTDR TQGSSIEEKR CAMTWHYRKA DPENGAFQAL ECEALLEELV CSKYDVEIMR 720
721 GKANLEVRPS SINKGGIVKQ ILSSYPEDSL PSFIFCAGDD RTDEDMFRSL HKNTRINKET 780
781 SFAVTIGSDK KLSIADWCIA DPANVIDILA DLANFTN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
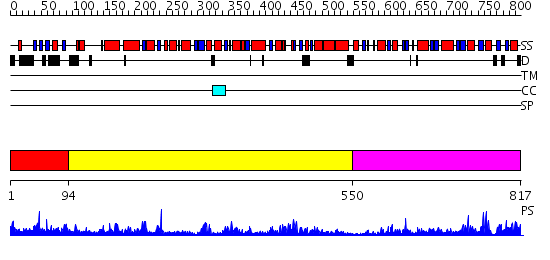
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [94..549] | 68.39794 | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. | |
| 3 | View Details | [550..817] | 30.39794 | Crystal structure of NYSGRC target T1436: A Hypothetical protein yidA. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)