













| General Information: |
|
| Name(s) found: |
tnr3 /
SPAC6F12.05c
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 569 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA cytosol [IDA |
| Biological Process: |
thiamin diphosphate biosynthetic process
[IC]
negative regulation of thiamin biosynthetic process [IMP |
| Molecular Function: |
thiamin diphosphokinase activity
[IGI ATP binding [IEA] hydrolase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMSSITSAK QLNAEELLDE CDSFNGEFVP GTIPFRANGA AIGYVTPLVL EILIKADNFK 60
61 FNWVYVPGEY IEINASTFEK RTDILAKVLE HWRHNNTFGI ADQWRNELYT VYGKSKKPVL 120
121 AVERGGFWLF GFLSTGVHCT MYIPATKEHP LRIWVPRRSP TKQTWPNYLD NSVAGGIAHG 180
181 DSVIGTMIKE FSEEANLDVS SMNLIPCGTV SYIKMEKRHW IQPELQYVFD LPVDDLVIPR 240
241 INDGEVAGFS LLPLNQVLHE LELKSFKPNC ALVLLDFLIR HGIITPQHPQ YLQTLERIHR 300
301 PLPVPVGKYE RGDSFEDTSK KAETCVPAKP QKATHQLAPC KAWLRDYDTD QKFAVLLLNQ 360
361 PIDIPDDRFR TLWKRASIRV CADGGANQLR NYDSSLKPDY VVGDFDSLTD ETKAYYKEMG 420
421 VNIVFDPCQN TTDFMKCHKI IKEHGIDTIF VLCGMGGRVD HAIGNLNHLF WAASISEKNE 480
481 VFLLTELNVS TLLQPGINHV DCHDNIGLHC GLLPVGQSVY VKKTSGLEWN IEDRICQFGG 540
541 LVSSCNVVTK ATVTIEVNNF IVWTMETRL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
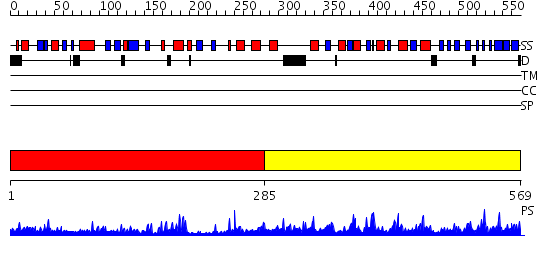
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..284] | 18.30103 | Crystal structure of NADH pyrophosphatase (1790429) from Escherichia coli k12 at 2.20 A resolution | |
| 2 | View Details | [285..569] | 70.522879 | Thiamin pyrophosphokinase, substrate-binding domain; Thiamin pyrophosphokinase, catalytic domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)