













| General Information: |
|
| Name(s) found: |
sid4 /
SPBC244.01c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 660 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA spindle pole body [IDA |
| Biological Process: |
barrier septum formation
[IMP cytokinesis [IMP septation initiation signaling [IMP cellular protein localization [IGI |
| Molecular Function: |
protein binding
[IPI structural molecule activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDEAFGDSLS TDYRWLGHSH FDSHPSAGDS IYFDSLDEDA DPSRTARIDR IAELLDGLND 60
61 EQISELVNGV NSTTIKENTK KEISNPNDST RLPLEQIPGS NNLFLDPRHQ LGDNSQNAQR 120
121 HHEPSFNSEK ASYTSTPYKN VAPKVIDSPS ARHMHSNSPS FPPSQSHTSS YDQSPKGQLR 180
181 DNISVPNQQD GLDPEVFNQQ SKETKKSLQV PPSRNVPPPV TRPNQYNPEP NFSLSSGYPQ 240
241 QHFSQPELQN RNVHLETVPE SYPVPPSGYP LTSSTCVSSI SQPIQSTDCQ KAQENLSNNK 300
301 QMSSNDQDID PFKQAITDLP PSFVNIVLEM NATIQSLSNQ CQQRDKQIEN ITKQLLMNQQ 360
361 DYCPTTMSTT VSTPLCPPKR FPKSTKDFKE QKPDTKQVRS ATISNDFNLK GNGRYNEKSQ 420
421 IAVPSEIRVQ LSTLDAILLQ FEHLRKELTQ ARKEIQILRH TSQNGDQESN ESSKNAITTK 480
481 TTDKGNNKEN TMLNDGSTAP AKNDIRNVIN TNNLDAKLSD ESELMIEKNK SYSTPASSTI 540
541 PTFHTSQPLT SLNMPDSRFN LAKEKQLYYR LGLQHIDQQC SVETANMLKT VLVQLNIPFA 600
601 IFPSTIGQVR RQLQQGRRLY QWARNIHYLI YEENMRDGLV SKQCLADMLK KIRELKKRSL 660
661 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
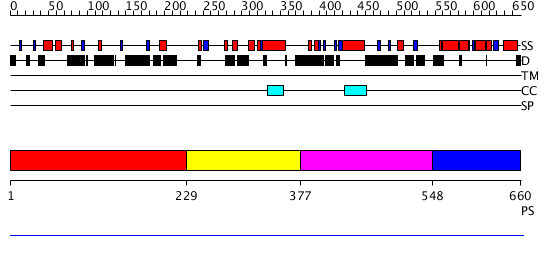
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..228] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [229..376] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [377..547] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [548..660] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)