













| General Information: |
|
| Name(s) found: |
pob1 /
SPBC1289.04c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 871 amino acids |
Gene Ontology: |
|
| Cellular Component: |
medial cortex
[IDA cell division site [IDA cell tip [IDA cell cortex of cell tip [IDA extrinsic to membrane [IEA] |
| Biological Process: |
unidimensional cell growth
[IMP establishment or maintenance of cell polarity [IMP cytokinetic cell separation [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASQRFVIAL HSFPGKSSDE LPLVEGRKYL LIKMDEEFGD GWWEGEDEQG NRGIFPASHV 60
61 ELISDERSDS SDSRRGKEDF SISTAEVTRS SLSSSRSTSS RSDKDSEKLY SNNSLSSSHS 120
121 SILNGPLDSL SKPSVPSNFN SMFPSSKQEG PSPLLDNQPS SDLSNFNTID ADYNNASAST 180
181 SAPATSASLK KVLSAEDSVR ETITDIETAL QNMSTSASRT PNDSSPLPYI ENRPASSLAV 240
241 SEKIQNVPNW STEEVVEWLM NAGLGSVAPN FAENEITGEI LLGLDSNVLK ELNITSFGKR 300
301 FEVLRKIQQL KDSYEQSLYE EYPQFAEPIS VSQSSDSSSS IPKKSNDEAG GSPSKSSPTR 360
361 PGFNDYVNRP TSVMPSLSNM IVSPDLDSSP STDWNQYVIP PLATPSSRNS KSTQSAVPEN 420
421 VSRFDSNEPS ATSPILKRSS PTDSISQNSG LPSRLTEPIS SPSTSSIDVD KEGTSFPGLP 480
481 YHSSKGNLYA PQPSSNVPTK FTGGASESSS VPPRPIPSAM KGKAPASAIS IEALEELDPP 540
541 KITTIDGESP SSISSRLPSS NLEQGSSSSV TKSPESMPDP SAKASSPVTS KGVSINEKSA 600
601 VNNYATPLSK PQPKDTKGSK LGNTFVAPSP AASLPASPPV GTELKTRPTL RSVASSPLNK 660
661 EPIGKRKSKR DIFGRQKVLP TGISEGLSNI PAKEAIKTAD CHGWMRKRSD RYGVWKSRYF 720
721 VLKGTRLSYY HSLNDASEKG LIDMTSHRVT KTDDIVLSGG KTAIKLIPPA PGAAKAAVMF 780
781 TPPKVHYFTC ENNEELHRWS SAFLKATVER DMSVPVLTTS RMPTISLSKA KELRTRPPSL 840
841 LIDDENEANL TSSIGLKKNA KQKNKKSSKQ K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
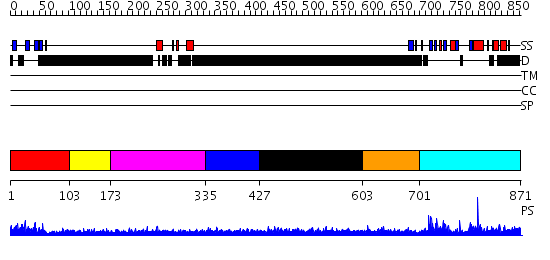
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | 9.30103 | No description for 2eyzA was found. | |
| 2 | View Details | [103..172] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [173..334] | 2.48 | RNA recognition by the Vts1 SAM domain | |
| 4 | View Details | [335..426] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [427..602] | 5.251991 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [603..700] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [701..871] | 36.69897 | Grp1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||
| 7 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)