













| General Information: |
|
| Name(s) found: |
pds5 /
SPAC110.02
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1205 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitotic cohesin complex
[IGI chromosome, centromeric region [IEA] condensed nuclear chromosome [IC |
| Biological Process: |
attachment of spindle microtubules to kinetochore involved in homologous chromosome segregation
[IMP sister chromatid cohesion [IGI chromosome segregation [IMP response to DNA damage stimulus [IMP |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIKLQFQRNI VPTHDNPLTT SEILKRLRDL LGELTSLSQD TIDRDSVLPV ARSLVNNNLL 60
61 HHKDKGIRSY TLCCIVELLR LCAPDAPFTL SQLEDIFQVI LKILSGLMNQ ESTYYPQIYE 120
121 ILESLSNVKS AVLIVDLPNA EEFLVNIFRL FFDLARKGTT KNVEFYMLDI INQLINEINT 180
181 IPAAALNILF AQLISGKGVR QTIGSSDSTN HGPAFQLARN IFHDSADRLQ RYVCQYFSDI 240
241 IFDSRDSLSD SMTTPEFIFS HNLVLQLWKY APTTLLNIIP QFENELQAEQ TSVRLVAIET 300
301 VGLMLQDNAI WSDYPRVWSA FCGRLNDKSV ACRIKCIEVA SNALQNSLAT SEIIENVVQM 360
361 LQSKLADTDE KVRVATLKTI EQLTFETFKM QFSVQALKLM GDRLRDRKLN VRLQAIRTLS 420
421 QIYNRAYQDL IDGVEYSIQM FSWIPSSLLE VFYVNDETTN AAVEICMAEL VLQYLSSDTQ 480
481 TRLNRLFLSI KYFSEKAMRV FILLLQRQVK YSELLNYYIE CCKNYNGGVM DNDEESITNK 540
541 LKKVIDIISS KSSNPTLTEA TFRKFAELND RQSYKMLLQT FSIKSEYQVV LKSIKYLFKR 600
601 VSETLSTASL ECFRIFVYRS ALFAFNKSNV HEIIQLLNEP VKYHNFLKPS EALLQHLPLI 660
661 HPNIYGEVVI EVENIIVSSG IESDPKVIKA LSQFSKRKKN FSIQTTTAEI LRKLCLHGTQ 720
721 EQAKQAATII AITETKEFKL DMITNIVENL EYNGGLPVRL MTLGQLFLYT LEEVEKVADQ 780
781 VTEFLVKKVI QRFPEKYDDT HNDEEWCTYE KLDNLTMCKV LAIRVLVNRL RAAAGGTEAL 840
841 NIGAPIIKLL KVLLMADGEL SPFKNTPKIS RAYLRLTASK YFLKLCSIPF YAEHIDFSSY 900
901 VQISLLCQDE NFDVRNLFLT KLQKQLQLKK LPISYYPLLF LTAVDPEEEI KTKASIWIRS 960
961 QVAFFQKTHD FTMEYVATYL IHLLSHHPDI SSIESENSLD FIAYIRFYVD TVVNSENVPI1020
1021 VFHLMQRIKQ SYDVIEDGNN YIYVLSDMAQ KILQVKSQNF GWSLTTYPKQ IKLPYEILRP1080
1081 IPSIDEKKRI FNKIFITPKM ESQIEHAIRT PVSSFAKQTT NKHANLKQKK THSSKSDKKS1140
1141 SRRRKNEKRR KLNEQNPNIR NVPERSSSRF QGIRINYSEA PSSSEEISEE EEEISEEDFD1200
1201 EIEDL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Unpublished Data |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
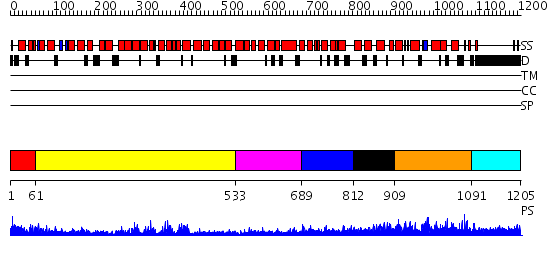
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..60] | 67.154902 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 2 | View Details | [61..532] | 28.69897 | Crystal Structure of The Cand1-Cul1-Roc1 Complex | |
| 3 | View Details | [533..688] | 19.0 | Structure of the Kap60p:Nup2 complex | |
| 4 | View Details | [689..811] | 76.69897 | AP1 clathrin adaptor core | |
| 5 | View Details | [812..908] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [909..1090] | 2.059949 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1091..1205] | 3.044988 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)