













| General Information: |
|
| Name(s) found: |
mex67 /
SPBC1921.03c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 596 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear membrane [IDA nuclear RNA export factor complex [IPI spindle [IDA cytosol [IDA nucleolus [IDA nuclear pore [ISS |
| Biological Process: |
ribosomal large subunit export from nucleus
[ISS]
poly(A)+ mRNA export from nucleus [IGI |
| Molecular Function: |
protein binding
[IPI protein heterodimerization activity [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRRKRERRN AVKENEMVID TPLEKRRTPG KPRATREPPI SVVITGHSKG SEDDLISFVW 60
61 RKVKVRLMNI SYSPASVTAV VKSQDFSRLN GLNGAAFAGD HLAIRRVDGA SNVTQDYRKA 120
121 KTKRSFRSVS APSLSALATQ AQRNVSKTLP QSTNETIEKL RQFLQTRYQP ATKFLDLGNL 180
181 QQDPLLKQMG ILAEASTKSK MFPALMKVAS LNFPDVISVS LSDNNLQSVT AVTTLAQTWP 240
241 KLLNLSLANN RITSLSDLDP WSPKTKLPEL QELVLVGNPI VTTFANRAMD YQREMVSRFP 300
301 KLRLLDGNSI NSEIIASQST VPFPVYQSFF DKVETEQIVN SFLAAFFKGW DENRSALVNQ 360
361 LYSPNATFSI SLNASNVRTN FSQKTDTKKW GAYKMKSRNL LYSQSQKESK SRLFNGHEEI 420
421 SNAVKSLPAT AHDLSDRSQW VFDGWNLVLP SVGAAIKIVV HGQFEEPQNK RLLRSFDRTL 480
481 LILPGGSTGI LIINDLLVIR SFAGSLGWLP GQSSVRTSNN AMSASASKPS DIVQPRPEQA 540
541 MLDTRQQIVL KIKAETGLND YYAHMCCEQN NWDYNSALAS FLELKSRNVI PAEAFS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
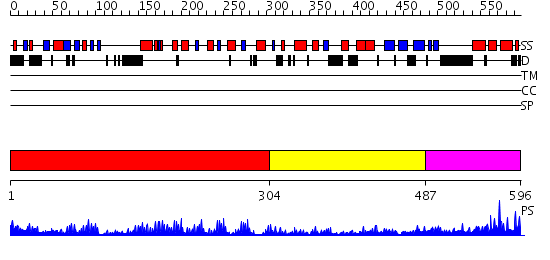
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..303] | 5.30103 | Crystal structure of CD14 | |
| 2 | View Details | [304..486] | 11.154902 | Crystal structure of the C. albicans Mtr2-Mex67 M domain complex | |
| 3 | View Details | [487..596] | 46.522879 | NTF2-like domain of Tip associating protein, TAP |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.45 |
Source: Reynolds et al. (2008)