













| General Information: |
|
| Name(s) found: |
met9 /
SPAC56F8.10
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 603 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [ISS cytosol [IDA |
| Biological Process: |
methionine biosynthetic process
[IDA |
| Molecular Function: |
methylenetetrahydrofolate reductase (NADPH) activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKISDKLLHP DWKEKVTYSY EFFPPKTSTG VQNLYNRIDR MKTWGRPMFV DVTWGAGGTS 60
61 SELTPGIVNV IQTDFEVDTC MHLTCTNMST EMIDAALKRA HETGCRNILA LRGDPVKDTD 120
121 WTEGESGFRY ASDLVRYIRT HYNDEFCIGV AGYPEGYSPD DDIDESIKHL KLKVDEGADF 180
181 IVTQMFYDVD NFIAWVDKVR AAGINIPIFP GIMPIQAWDS FIRRAKWSGV KIPQHFMDTL 240
241 VPVKDDDEGV RERGVELIVE MCRKLIASGI TRLHFYTMNL EKAVKMIIER LGLLDENLAP 300
301 IVDTNNVELT NASSQDRRIN EGVRPIFWRT RNESYVSRTD QWDELPHGRW GDSRSPAFGE 360
361 FDAIRYGLRM SPKEITTSWG SPKSYSEIGD LFARYCEKKI SSLPWSDLPI SDEADLIRDQ 420
421 LLSMNRNAFL TINSQPALNG EKSSHPVFGW GPPNGYVFQK PYVEFFVHPS LLNELKETVK 480
481 KLNSVSYFVT NKNGDLDTNS QYEIPNAVTW GVFPNREIIQ PTIVESTSFL AWKDEAYSLG 540
541 MEWANAYSPD SISRKLLVSM MKEWFLCVIV DNDFQNGQSL FDVFNKMRSL KDIHPELYYA 600
601 NAS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
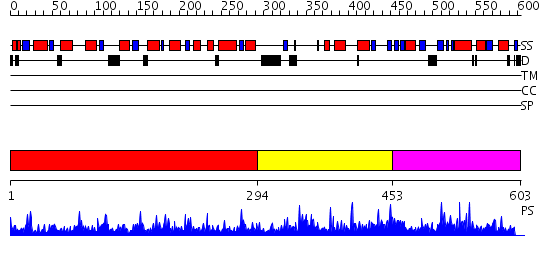
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..293] | 104.0 | 5,10-Methylenetetrahydrofolate Reductase from Thermus thermophilus HB8 | |
| 2 | View Details | [294..452] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [453..603] | 2.026951 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)