













| General Information: |
|
| Name(s) found: |
meu6 /
SPBC428.07
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 651 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nucleolus [IDA |
| Biological Process: |
meiotic chromosome segregation
[IMP meiosis [IEP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSYEGREERP EQIAEPTFEN VSEHNEHDSG SAEAQVETLE VPLSNETDND DATENAVPAE 60
61 VQAPEEPKEP ETVDNIDPAD DDPNSVAAPK VEEKKSKKKA KDEKPLTYTT GGYLYCRSNG 120
121 LIPHVMKRYF YMLEGPMSMD LLDHYYKKHF HGTLQGNPTE PSLASEQMHD SDADSLFQSA 180
181 NDHLNRIAKA TQDCRGLLFY SKSQSTSVPS GIINLTDAVS IEPGTGNKFT INFNNGKSET 240
241 LEALDPESKN TWITDLKNAI AKKKETKDKI KELRAYNDTL KRLGKLAARS GPSASETFRY 300
301 FLPMLSNGRD AKKSSSKSHG GKQNNSVAKD GLFDFFQNMI KGKTPQNDTA SPIDNETSAD 360
361 PVDTTVEAQS VEVPENETNQ IPTTEEHFPA TTEEVAPASE EKPATGPAEE STSTQNVEQA 420
421 STQNDNGTPI GQIADAIEED VKGTAQSVEQ AFIEETDIAL PDIALPDVAT PTADDQDPST 480
481 AATNEESVVS NEDRTSKKAG KKHHRHHKKK KGGNKRVFGF KLHKYAKPTK SVKTLQADPS 540
541 LEFAKAGFEI FPSNLSKKLS GLVQKKVHKK FDKDGRLKEI SQFSKTTIKP ETPLTPTTTP 600
601 TPRTAAQEDP AEETTDALAS AEGENNQMPQ VLETEAAPSI ASEGRSITVN A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
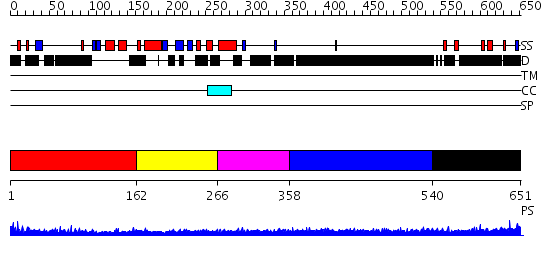
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [162..265] | 0.959 | No description for 2dhkA was found. | |
| 3 | View Details | [266..357] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [358..539] | 4.214996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [540..651] | 2.199993 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)