













| General Information: |
|
| Name(s) found: |
cgs2 /
SPCC285.09c
[Sanger Pombe]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 346 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
negative regulation of adenylate cyclase activity
[TAS cAMP catabolic process [ISS positive regulation of gene-specific transcription from RNA polymerase II promoter [IMP cAMP-mediated signaling [IMP regulation of conjugation with cellular fusion [IMP cellular response to nitrogen starvation [IMP |
| Molecular Function: |
3',5'-cyclic-AMP phosphodiesterase activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHAALEIKEA EFQTDQVVGL VENSFTLYSL GQNGGPLESC CSSHLISDGA FQEIISLDGG 60
61 SHLSALVELI QSKHLSVDSW SSITKYDNYT VENESYAKAW HLSEQRIKTF LITHCHLDHI 120
121 YGAVINSAMF GPQNPRTIVG LNYVIDTLKK HVFNNLLWPS LDKAGFINFQ VVEPSMYTSL 180
181 TTTLSILPFP VNHGSSFGQE LKSSAFLFRN NLSDRYFLAF GDVEPDMVAS EPLNIHIWRA 240
241 CSSLIAQRKL SHILIECSTP DIPDTLLFGH FCPRHLVNEL CILQSLVQSY GVIMPTLTCL 300
301 LTHLKSHPLQ SANPADVILE QLESLSSKSS LSVTFKILQR GQFYKF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
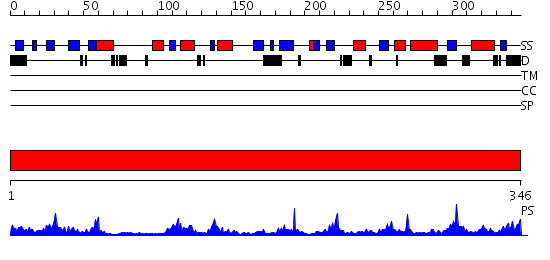
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..346] | 32.09691 | Crystal structure of RNase Z |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)