













| General Information: |
|
| Name(s) found: |
cdc45 /
SPAC17D4.02
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 638 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IDA DNA replication preinitiation complex [IDA nuclear replication fork [IC cytosol [IDA |
| Biological Process: |
DNA replication preinitiation complex assembly
[IMP DNA replication initiation [IC DNA strand elongation involved in DNA replication [TAS |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFIKRSDYAS AYLKIKEASV SGGCTVQLFV ALDPDALCAC KLLSTLLKGD FISHKIRPVS 60
61 GYRDLEQANK TLLEQNEDIK FIILLNCGTM VDLNNYLVSM EDVSIYVIDS HRPHNLNNIY 120
121 IENNIFVFDD GDIEEDMNKI HDAWYAFNSH ELSDEENSDS SNEREEEVED DNRSVESYSS 180
181 SDYQARSRRR FSEETTQRRA EIKEKRKKRK EFASILSEYY EKGSWYGESI TNILFAVASM 240
241 LGREDNDMLW LAIVGLTCLE IHCQSSKKYF NRSYSLLKDE VNRLNPSPLE NQIVGRAHGK 300
301 TPHDQSIRLE DEFRFMLVRH WSLYDSMLHS AYVGSRLHIW SEEGRKRLHK LLAKMGLSLV 360
361 ECKQTYIHMN MDLKKTLKSS LKRFAPFYGL DDVIFHSFTR TYGFKCTLSA SDVSYAISAL 420
421 LEMGNTGVLL QSKTVARSPD MTEEEYLEKF ENAQNQEWLH NFYDAYDALD DVDSLERALK 480
481 LAMHLQRAIV RTGITLLEKR AIKTLRSFRF GLINEGPDLK IFMHPLALTK MSLWIAEAIN 540
541 EQEREFGKLR HLPLVLAAFV EEKNRYLIVG TSTSAFTSNE DDDDDDGHGH NRFGVAFQEV 600
601 ANMTSATLQM DCFEASVIEC QKSDLGVFLE SLSFKTLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
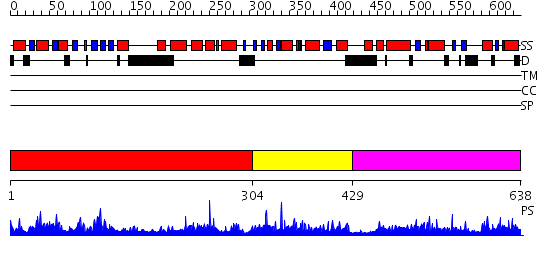
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..303] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [304..428] | 1.033972 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [429..638] | 1.031955 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)