













| General Information: |
|
| Name(s) found: |
ada1 /
SPBC106.04
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 846 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[ISS cytosol [IDA |
| Biological Process: |
IMP biosynthetic process
[ISS cellular nitrogen compound metabolic process [IC purine base metabolic process [IEA] |
| Molecular Function: |
AMP deaminase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVRPLSLCG LLPAEMNMEQ EDDQVPAVAA ETVPLKRYVT NPGANRDEEV AAAPSSQDTP 60
61 YFDYAYERSL RHQDAKFLAM NGTQNGRDGL PSKSPRRPSV SASTVRNSDD VNHSKAGPGS 120
121 GKLLNDTLQS KISSIHMPHV QQGDNAVVSS VGGPETDPGN METTDPLFSD ELAEIYLSIH 180
181 KCMDMRHKYI RVSLQGELDN PIDDDSWIIY PDCKEGEDDT GLFNFADCKI PGIENEMEYH 240
241 MDHQGIFQVY ENDSAYIAGT PSFHIPTIRD YYIDLEFLLS ASSDGPSKSF SFRRLQYLEG 300
301 RWNMYMLLNE YQELADTKKV PHRDFYNVRK VDTHVHHSAL ANQKHLLRFI KAKLRKCPNE 360
361 KVIWRDGKFL TLQEVFDSLK LTSYDLSIDT LDMHAHTDTF HRFDKFNLKY NPIGESRLRT 420
421 IFLKTDNDIN GRYLAELTKE VFTDLRTQKY QMAEYRISIY GRNREEWDKL AAWIIDNELF 480
481 SPNVRWLIQV PRLYDVYKKS GIVETFEEVV RNVFEPLFEV TKDPRTHPKL HVFLQRVIGF 540
541 DSVDDESKPE RRTFRKFPYP KHWDINLNPP YSYWLYYMYA NMTSLNSWRK IRGFNTFVLR 600
601 PHCGEAGDTD HLASAFLLSH GINHGILLRK VPFLQYLWYL DQIPIAMSPL SNNALFLAYD 660
661 KNPFLTYFKR GLNVSLSTDD PLQFAFTREP LIEEYAVAAQ IYKLSAVDMC ELARNSVLQS 720
721 GFERQLKERW LGVDFQDIDR TNVPIIRLAY RALTLTQEIA LVNKHVQPSK HPSNHDLEEL 780
781 IHKYDAMTGT SDPLSASPRT NDATISSRLS LHDGHDHGAF FPGLSVISER RRRKDSMASS 840
841 SQDLKD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
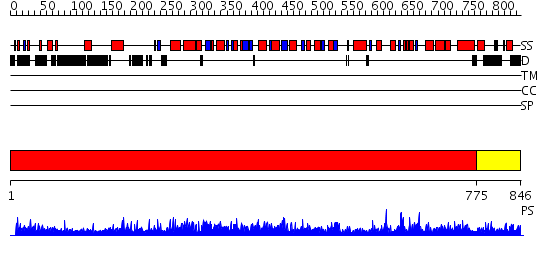
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..774] | 1000.0 | X-Ray Structure of Adenosine 5'-Monophosphate Deaminase from Arabidopsis Thaliana in Complex with Coformycin 5'-Phosphate | |
| 2 | View Details | [775..846] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.51 |
Source: Reynolds et al. (2008)