













| General Information: |
|
| Name(s) found: |
par2 /
SPAC6F12.12
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 627 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cell tip [IDA protein phosphatase type 2A complex [TAS |
| Biological Process: |
regulation of catalytic activity
[IC cellular response to stress [IMP regulation of septation initiation signaling [IMP |
| Molecular Function: |
protein phosphatase type 2A regulator activity
[IEA]
protein phosphatase regulator activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKGLRSKFVK ALSLKDEQGS HKNGHSKSHY ISKNGSYVET DDVKHTDTHH SSKHELKKLK 60
61 SHFLKDTLKH KRNHHANSNN EKHENSDKKI HTTVLASGHE DSDYSTFLPV IETSKVKDAN 120
121 HFPPNYPEPK NDSVSSNIDE FPNDSISSAS FLSVPQSTPP YLVSQPTPLN KFLAGAENVD 180
181 IPHSLRPVPR REHSSQFQVS EKRTLVRLPS FDDVHTSERE ELFIKKLEQC NIIFDFNDPS 240
241 SDLASKEIKR EALLQMIDYV SENRGISSAS LFPYVVNTFS LNVFRPISPA LNDYSSDMFA 300
301 LDDEPFLEPA WPHLEEVYLL FIKFLESPDF RASKAKSLVD RRFFNRLLAL FDTEDPRERE 360
361 LLKTTLHRIY GKFLNLRSYI RKSMNNVFLQ FIYEREKFHG IAELLEILGS IINGFAVPLK 420
421 EEHKIFLSKV LIPLHQTKSV FLYHPQLTYC IVQFIDKDPS LTKAVLTGIL KYWPRINSFK 480
481 ELLFLNEIED IFEVLEPSEF VNIMSPLFQQ LARSISSMHF QVAERALCLW SNEYFTSLVS 540
541 QNVVTLLPII YPSLYKTANE HWNSTIQAIA CNVLQIFVDM DADFFNGLVE DYKQAIIKQE 600
601 EVMIIRKQQW CQIEALAAEN KPTDYLR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
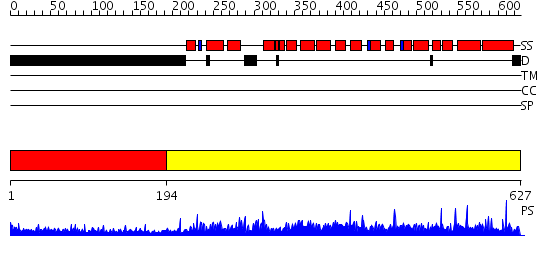
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..193] | 11.226997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [194..627] | 1000.0 | No description for 2nppB was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)