













| General Information: |
|
| Name(s) found: |
par1 /
SPCC188.02
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 548 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA spindle pole body [IDA cytosol [IDA protein phosphatase type 2A complex [ISS |
| Biological Process: |
negative regulation of septation initiation signaling
[IGI regulation of catalytic activity [IC] cellular response to stress [IMP regulation of septation initiation signaling [IMP |
| Molecular Function: |
protein phosphatase type 2A regulator activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKGIKSKMLS RGKSQDTQKS SKKKESKKSN SHDSSKAPKE SPSTDPNGSV IGAQNDFLTV 60
61 PKHSGKKVPI DTTPTPRDEI LLENVRTVRK QRSSLYHISE NRNLVRLPSF TDVPVNKWHS 120
121 LALEKLEQCC VVFDFNDPST DLYGKEVKRE ALQDLIDLIS VRKEAIDESL YPSIVHMFAV 180
181 NVFRPLPPPS NPPGEIMDLE EDEPALEVAW PHLHLVYDFF LRFFESPSLN TSVAKVYINQ 240
241 KFIRKLLVLF DSEDPRERDF LKTTLHRIYG KFLSLRAFIR RSINNLFLQF VYENEQFNGI 300
301 AELLEILGSI INGFALPLKE EHKIFLSRVL IPLHKAKSLP LYYPQIAYGI VQFVEKDSSV 360
361 TEEVVLGLLR YWPKVNSSKE VLFLNEIEDI IEVMEPSEFL KIQVPLFHKL ATSISSQNFQ 420
421 VAERALYFFN NDYFVHLVEE NVDIILPIIY PALFEISKSH WNRVIHSMVC NVLKLFMDIN 480
481 PSLFDEVDAE YSESRRKKED EEIIREERWT ILENIAKENA MKLKSQNPTT VHSTTERLKK 540
541 LSLDYTNG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
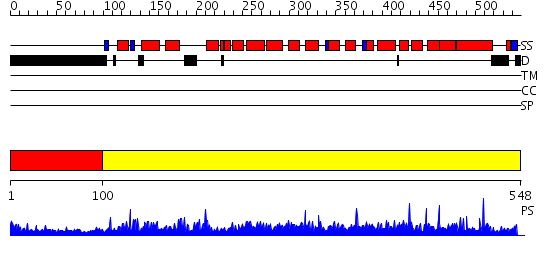
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [100..548] | 1000.0 | No description for 2nppB was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)