













| General Information: |
|
| Name(s) found: |
slgA-PD /
FBpp0076903
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 681 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[ISS][NAS]
|
| Biological Process: |
locomotory behavior
[IMP]
glutamate biosynthetic process [ISS][NAS] phototaxis [IMP] oxidation reduction [IEA] proline catabolic process [ISS][NAS] |
| Molecular Function: |
proline dehydrogenase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALLRSLSAQ RTAISLVYGR NSSKSSNSVA VAACRSFHQR GSGSTSIAGE GAASESTRGV 60
61 NGARFLHSGD RPLQASTLVQ PEVVSSETVK RSMKQESSQE KNPSPAGSPQ RDPLDVSFND 120
121 PIAAFKSKTT GELIRAYLVY MICSSEKLVE HNMTLMKLAR NLLGQKLFVL LMKSSFYGHF 180
181 VAGENRHTIV PALERLRSFG VKPILDYSVE EDITQEEAEK REVESSVSSA GDKKEEGSMP 240
241 QYHVDKSFAD RRYKVSSART YFYLNEATCE RNMEIFIKCL EAVSDDDRKA PRAVATGATF 300
301 GTGITAIKLT ALGRPQLLLQ LSEVIMRTRK YMEDMVGGQG NVLTHHKTIK DLEKYYATLG 360
361 DNKDVKEFLN NVTSDKEGIL HLFPWSGIVD EDSQLSDTFR VPDPQTGQMR RLISQIPPKE 420
421 EEMFRNMIRR LNTIVKAAAD LDVRIMVDAE QTYFQPAISR ITLEMMRKYN KDKAIVFNTY 480
481 QCYLRETFRE VNTDLEQAKR QNFYFGAKLV RGAYMDQERD RAKSLGYPDP VNPTFEATTD 540
541 MYHRTLSECL RRIKLMKDCD DDARKIGIMV ASHNEDTVRF AIQQMKEIGI SPEDKVICFG 600
601 QLLGMCDYIT FPLGQAGYSA YKYIPYGPVE EVLPYLSRRA QENKGVLKKI KKEKRLLLSE 660
661 IRRRLMRGQL FYKPKGNYVP I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
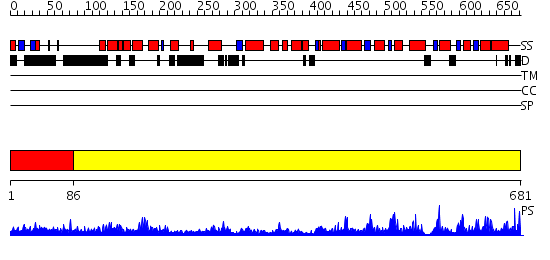
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [86..681] | 123.0 | N-terminal domain of bifunctional PutA protein; Proline dehydrohenase domain of bifunctional PutA protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)