













| General Information: |
|
| Name(s) found: |
CG1486-PB /
FBpp0076892
[FlyBase]
CG1486-PA / FBpp0076891 [FlyBase] |
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 852 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
carboxylic acid metabolic process
[IEA]
|
| Molecular Function: |
pyridoxal phosphate binding
[IEA]
carboxy-lyase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAAGRSVDD PGQSPLVDQE PDPGQSTPAV AEIAASSIRS GLAELELRSS QVLQRLENVK 60
61 VSTTPESPNP VENEEEAGER VTSEDLLPAK HRVPSDILKS LEQLVSYTDS DDDPEFPLPA 120
121 LDDVSHLALI SHSIVAYLSH LDRQQLLRVT NSISGDATRW LGTLFHFAHP ASSFHADNAD 180
181 AVLRTVRLAI VARCPGYLEG GIPALAQPTF YISENTTPMR LHYACRQLGI PLEAIKVIPE 240
241 HSQSGTMDVT LLQKQIQQDV GNNRTPLLVV ADIGASLCGY VDNLLRLRDV CKAHNMWLHA 300
301 SGHGLAALVC AQNQGHVEEV LHSMALNLGS WLGVPSLPIV LLHRPLQNSA LSAFESDPIL 360
361 SRRLNALSLW TSLQALGRKA IAERLHVAFQ TCSILFEIAS KCEGIRVLSH TPGAQTGASL 420
421 SDVIQSPFDV QALFDAAAPV VAYQFDGSTT IPLGGSGSSA AAAAAERETA EGLKPLEKIN 480
481 NASYFDRLNS WLGQILQRDC PNFDFEVIEH PTHGSCIRYC PLELGLGEQP PSSENLESFA 540
541 QSLEAHVDIL RATIKHKARF IHLVECSDVL RLVPLPEWAG MGGVRFVPEG WESLLTDQAK 600
601 TELNKLNIDL VEALKSTDNA FSLGEGTDGL ICVRFGMVTH ETEVEELLDL VVTVGKSVQE 660
661 NSRVLDTMSE IVKKGIEAVT ADLQRESEEK LWQEGILRHV PVVGRVFNWW SPPAKESGIK 720
721 GRSLNLTQGV VESTENIYKY HMQMTGATAH QLPANRSPPT PMVQTPVVAP ALPPVFPTVE 780
781 PVPGNGNGSG ASEGTPVQSG EASGSMPAAS GAAPSAAPTQ NHVDHARTVS QSSAASSTVP 840
841 ELVAASSAIN NN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
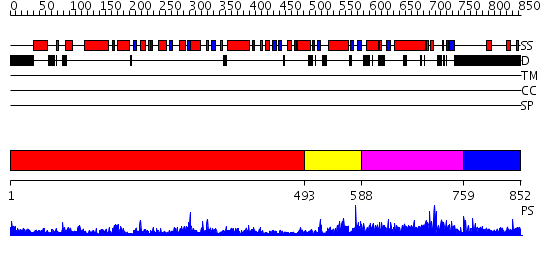
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..492] | 82.154902 | No description for 2jisA was found. | |
| 2 | View Details | [493..587] | 86.0 | Cysteine desulfurase IscS | |
| 3 | View Details | [588..758] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [759..852] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)