













| General Information: |
|
| Name(s) found: |
FBpp0310187
[FlyBase]
Grip84-PC / FBpp0074546 [FlyBase] |
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 819 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYALLVFVER YSECKPVSES SSSTSLSAMG LPHGNKTSDV NSAAGSVPTT LAIASTSTIL 60
61 TTSQNVSGST RLSLTQSQDF PTSTPVNCKK ATESDTTPVV FVRRGPMDRN RGAGKDERND 120
121 LSVIKERVLN AVSDQSLSGY RSVTNKTGNS PKSMPNDLVT LTDVPDEYRT HLLWEYYKVD 180
181 GDKVPRAEIA AMPLLSQESM LLDELLHCLT GIRESLLVPQ KPIISAVGLA KYDTDFDIHT 240
241 HLDRSLTHQV REILPLASYF MGVQKIIAAT DGLGQVMNSL NEALQELTHD FYLIIVQAEQ 300
301 ELRHNRLTLQ KLLYYLQPTM WVMHEVWSSL VIIQLSDSRD AEVLTYLHER IKRLEGNKDA 360
361 QQLIIGLVRK AAKPYMRMLQ MWIQKGVIVD RHREFLVVDN EVIHRDELPE HYSDDYWERR 420
421 YTLRDEQIPS FLAKYSDKIL RTGKYLNVIR QCGKRVMPTQ EMNLEFDPTS ERHVSVINDA 480
481 YYFAARMLLD VLLTENDLMG HLQSVKRYLL LNQGDFTMQF MDACEDELTK NVDHVLPMTL 540
541 ENLLGLTLRI SSARNDPYKD DLHCELLPYD LVTQMSKIMK KEENWQAQPR LDLSGLECFA 600
601 FTYEVKWPCS LVLNHISISK YQMLFRQLFY CKHVERQLCK IWKENSIARQ FEPQAASLYR 660
661 AAFTLRQRMM NAIQNLEYYM MIEIIEPNWH IFIEKMKTVE NVDNVLRLHQ DFLDSCLKNC 720
721 MLTESSHLNR SIFKLCKICL KYCEFIQIET PLDPTDTFSE RVRRFDLEFT QLLISFLKQI 780
781 NSMAKKNTAD CFMNLVHRIN FNAFYTDQMD KMCVEDAIG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
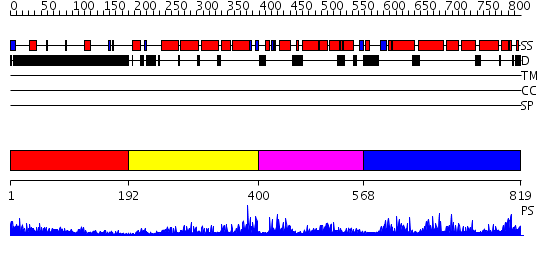
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..191] | 2.01 | No description for 2fj7I was found. | |
| 2 | View Details | [192..399] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [400..567] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [568..819] | 1.18997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)