













| General Information: |
|
| Name(s) found: |
rictor-PA /
FBpp0074458
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1936 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to DNA damage stimulus
[IMP]
|
| Molecular Function: |
binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASQHSSWRF GKRSKLQLRI KVSQDPEDFY RLDPQRSAAE NAFEIYSMLC LEETRDTKRL 60
61 FLLNALASLC LGARKGSHHS NIRFTTEELL YCLSASLVHT FTQVRAAALR TIRYAISTPN 120
121 DIKTFNALQL QHLLCRSIDL MLKNDDERVQ ALKLVRKMLA IAPEDISPAV VRCLVSLADS 180
181 GIEENDNLLR ACLATLAEFA VLNPALLIVC GGVTSITRNV LECHNPRIAE SLCGVLLYLL 240
241 EWPQTRNICG VRLDCLAAPY CDFTYRQSIV DKNKDARELR YTSCRLALLS VLRSWTGTLE 300
301 FCDPSKPSGL KAIVDALYLN QIEVRKAILD LLYELLTLPQ PTWTDDYAVA LQAVDPSDFQ 360
361 DTWLLSNGFV SAEGRSILPT LAARAPSVVE QHLALMLYCF LETGLLNALV EVVVSSDQFV 420
421 SVQATVLIGK ILQLMHTHLP PDICCTSPAL PTLVSHATLG NQQANAAVAA LQNYQKLLRQ 480
481 RPASRSLFLD SIIQGGALIQ TRLFRRHLNV QEQAGPVLQL QETAEAAAFA VPPVTQFRGT 540
541 LDRSRLDSVS SSDESNSQAS TSSRSSFRLK RKFLPQALYD NFRAFNRLLT DSRVLSQADA 600
601 HLWDWDVITT ILKSNLIRKL DYTQGKFLKR LVDFYKPRNN RFSHQDLVPG QRLPTYVSAG 660
661 LDLIDVLLSS NELECMRFIT DYFSDISQQL AAVTTSNRAH DCLFSPQHMN NTMCQQYFLY 720
721 IGRMCRTVKG IEVLKNTTVF EYLINLVRVT DHVCYVKLIV SGLNYSYEKL PRQVLEKALT 780
781 SAKTRGRLYS TQFMAVLLRA RLPHFEVWGI PLIINQTRDS DRSVGLAAMD VLEEACHDKY 840
841 YLEEIVSRWP NLTHRGDAGR LLMARYYSLP RGLNSTMARI EDEIRYWRNG YNKRYVLLVE 900
901 ADTHSSLTLH IRNEDGYYSR RNCNQRPQTV PPNVAPHLYG QMAQTGQGMT ALRKHGDLPQ 960
961 LLELLRRAKC TDDAECLELK AAIWALAHAS THSNGIEYFV ELNARLYEKL IVLVTKCEVY1020
1021 SVRATCFSAL GLIAGTQAGA NILFKLNWLS VRHDKNTMWP VHQPEDWMSS QYTPVRHYYE1080
1081 DVPADNMTMM DDYIERFYET GSDFWHMLAD QTDASGGGGG VAGTVVGMHP IVQDPNATIT1140
1141 TTDSVRTTTD DVHPRMLVGV AAKSKTLPEG SNLRQGKHQR SLSESKTTDV ISLLGSGVGT1200
1201 MSGAVLYPTP YQHRIRYNSC TDSNTSGVSS CESVTGRTAA AYAAAANELQ QFPLSPIPSM1260
1261 SNLLESDELF RRNQLATSML PSTLSPMAMK GYVQLRSLRK HSRPVFSESS AEFYDFAEIL1320
1321 DTPEVQMRKL DWTHSHRRLK VRSLDRQLSD VYRRLSADEL NVLLPTLNAP KFLPPNDLKG1380
1381 PCYAGICLPK NVLDLFPTRN LSRTYVSRDI QDQDIVGINL LNTMLRPQCL NDSLTNEGGD1440
1441 ESSVISSLSD VSSASRRQTR QLQQGAKHSR SLCLHCARSP RQQRNDSGSH SNGGGGASLA1500
1501 PCELYSSAAA ALVAAGIQAG PVLAKKSGSS AQGASAALGA DISFHSPESM LSEDSLPDRL1560
1561 TASILYNVQR LANPVSAKQS KMALLELKQK HPHAFQDICL YSEACKTIGR SSYRMIARRF1620
1621 LQELFLDLNF DSFYVEPQLI IGARKFSAEE KAEATAPTAM QAMPFKQTIL KPKPAAQLAS1680
1681 VYETSWENLL MDQSPRINKS SPADQPANAQ TKAKARNQLH IDDTESSLSS TDGENDSGED1740
1741 VCDGSGAGTQ ASSVTTAHTA LPTGNSNRSS IGTCTASQLV PGAATSNSAG AGTSASDNAS1800
1801 TSLKNENRQF TRGRFYTLEL DLSCTKNKFP IKNRSQRTPT PPPIESLIES KPTVQTDVNT1860
1861 SALPIMQRTG GSNDSSSALR YSTVTKSLSA SMARPVGSLY CEKRLQSSKS EAVLDEVAKA1920
1921 RRASGGVGLT TVRDNS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
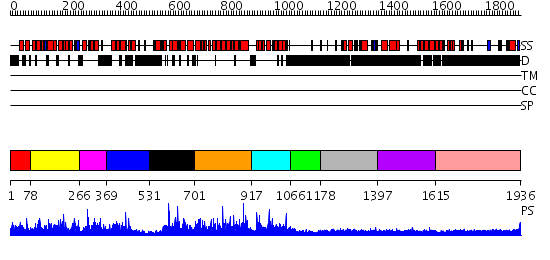
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [78..265] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [266..368] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [369..530] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [531..700] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [701..916] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [917..1065] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1066..1177] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1178..1396] | 1.162995 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1397..1614] | 2.174997 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1615..1936] | 2.24 | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)