













| General Information: |
|
| Name(s) found: |
FBpp0293027
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1509 amino acids |
Gene Ontology: |
|
| Cellular Component: |
filamentous actin
[IDA]
|
| Biological Process: |
bristle morphogenesis
[IMP]
epidermal cell differentiation [IMP] antennal morphogenesis [IMP] imaginal disc-derived wing hair organization [IMP] actin filament bundle assembly [IMP] cuticle pattern formation [IMP] |
| Molecular Function: |
actin binding
[ISS][NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTSLTSYRR SKQKSKSNND IHTIIATEVV TAAILSSSVH SSSTGSEIGD GGQGNTSLIE 60
61 EAAAQDQSSE EQDRQHQHAK DPLAQTEMNH HPHSHHQRKS AVEVVRQQMD DDGDQDSNDD 120
121 DETMPMMSGV GRKEQNNRRS VVSANTQMDN DVTPVYLAAQ EGHLEVLKFL VLEAGGSLYV 180
181 RARDGMAPIH AASQMGCLDC LKWMVQDQGV DPNLRDGDGA TPLHFAASRG HLSVVRWLLN 240
241 HGAKLSLDKY GKSPINDAAE NQQVECLNVL VQHGTSVDYN GKSSSQRHKS QQQLHQQHQQ 300
301 QQQQQQQQHL SSSCNSNSNS SKQTSRSNTI KSKSSSTLSS DVEPFYLHPP ALAGGSGGSG 360
361 MGLGMGMGMG MGMSMGLGIS RKNSDALYSQ QSRSSSEKLY NGSSSLGGNP GGNSSSGGGG 420
421 VGGGGGSGGA LLPNDGLYVN PMRNGGMYNT PSPNGSISGE SFFLHDPQDI IYNRVRDLFV 480
481 DSDCSSSVKQ HGKSHAHQLL QSKAGNAMTI QADVHSSSSG AGSGSDESVS ISSSFNSPKQ 540
541 QQQLRSQSFN RHGSSSNISA SGNIKNNNYK AHKGKPSNQN GGGDHDYEDI YLVREESRKQ 600
601 HQQQQQHQLQ QQQQLQHQAT QQQQQQQQQQ QQLQHKYNVG RSRSRDSGSH SRSASASSTR 660
661 STDIVLQYSN HHLNNKRNMN NNSNMNNSSS NIAQSSSSNN NNNSLLNRNK SHSIIGLHSS 720
721 KYESSLKDNY SAKNVNLKNQ LLNGGIKSDT YESVCPPEDV AERTKQTHKN SMIRNNLADA 780
781 SSNNNTSGSI NNISNIGNMN GGNQSSRNLK RVSSAPPMQN LAVVNGPPPP PLPPPLRAPV 840
841 QQQRNNLSVD QPNSMTGNNG IYKKNVFGPN QGPTNANGTG GAAPPPVPAP NPVATEAVDS 900
901 DSGLEVVEEP SLRPSELVRG NHNRTMSTIS ANKKAKLLNA GSTNGSSIAA SSDDSQSRYG 960
961 GSVHAANGSA ANGHFYGYSE GNKNQGSNGN GNGNGNGGGG NYHSHPNQPH YATGQPHQYT1020
1021 PSLYGGGSSA EDHYELTQQQ QQIYGETHLP SGQRTGGPNL VNKQLVLPFV PPSFPNKSQD1080
1081 GVTHLIKPSE YLKSISIDKR SCPSSARSTD TEDYMQIQIA QQQSHPHQHQ HHPHPHPHPH1140
1141 HPHQLHGMGE QPPLPPPPPP LPHGMLPPQP PMGPPMGVAG GQDATMSGKS HKPKKPHGSA1200
1201 PNSQDTATRK QHQPLSAISI QDLNSVQLRR TDTQKVPKPY QMPARSLSMQ CLTSSTETYL1260
1261 KSDLIAELKI SKDIPGIKKM KVEQQMASRM DSEHYMEITK QFSGNNYVDQ IYLQIPERDQ1320
1321 AGNAIPDWKR QMMAKKAAER AKKDFEERMA QEAESRRLSQ IPQWKRDLLA RREETENKLK1380
1381 AAIYTPKVEE NNRVADTWRL KNRAMSIDNI NINLAGMEQH YQQIQFQQKQ QQLQLGNKEN1440
1441 HVDPKGADQD QEHGVGIGSG VGNANSGVIG GQELIDPSEG SQGRAEEEED NIIPWRAQLR1500
1501 KTNSRLSLI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
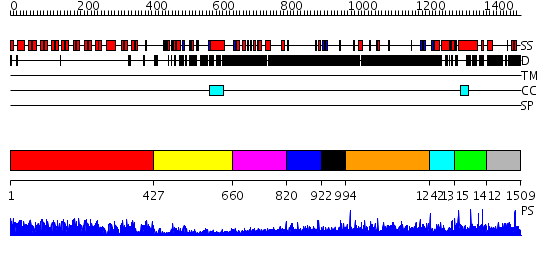
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..426] | 101.0 | Ankyrin-R | |
| 2 | View Details | [427..659] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [660..819] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [820..921] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [922..993] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [994..1241] | 2.105996 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1242..1314] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1315..1411] | 1.015991 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1412..1509] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.30 |
Source: Reynolds et al. (2008)