













| General Information: |
|
| Name(s) found: |
shtd-PA /
FBpp0073893
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2030 amino acids |
Gene Ontology: |
|
| Cellular Component: |
anaphase-promoting complex
[TAS]
|
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIAVAEPPLE FVPRGRQTSA EHPGPHDQPM ARHQLPTTEH LLLQRLQNVN ISSAGEDCPS 60
61 GGQESWTVRE LYDDYEASEE RAALRRKLTE RTKRCGLQAT SLPNRLLNPP LVRRVEPMCD 120
121 YMVNNEEELY VNRNTVVWTQ GVNDDDDTDN GVYRRMCFTT DTPVRFACFL NRSFVRGRLA 180
181 QLKAAVHPDD DHLTAICVMD QDALRVYCSN GEDFLANLDF PVSQLWQTKY GLLLEKDSSN 240
241 ALISHMSIPM PRLFSMSHPL HEACPVVLKT ATGSTGYMTE PEYTVVFTTE ESDLVMLYDA 300
301 KFFKHFVARL RKVTPEEINY VSQQMELGQT LMGPRSMAGN SFSSTKQTGA TPKATNLSFA 360
361 ARNINTTTTG MGNQFGLSQS QSFSGVLGQS NRASLGTPLS QLQSSISQQS MSVKDMRKLT 420
421 HVKPAKPIEP ELCMEHIWTE NIYGTQREFC EMATRAFIHT DLVGQTFLCY LLARSCRLQL 480
481 VRLTGYGRGE VQLSTHASTL AAKDAVGLKR MHMIAVLDPS GSLLLYTGTV LISKVHITPF 540
541 LAPTSIPTPL VTPMTAAPSP SASPAPSHVK TPMAAAGPAS GIPSGSSSFV EVRRSSLLPT 600
601 KAPGDVAAFE EELHMLSPIQ PQPVSYTQRQ AHNVCKSLRD PAGNRLTLVY ATGRMLRIAL 660
661 PFLNDTRLLT RCVATLRQVL SPTQFLDFVI RWYSDRNPPG SRNYSIEQEW LLFRSTLLAL 720
721 MGLTAAPDVD AGENYARCAT PPLHTQFGGG ATATESSDGS SCSSNSTLGG QDEPKKRRIY 780
781 NDCDDFTDDD WEFLLLQTTL APCGADGHSY SVNIGALLFR MIPAIFFSLH LLYEDLKLDA 840
841 DFYGALPYLA TFLHQLAIDM QLESYVLHYI LDFPELSNRT GKLSLLGAEH GAMMLHQELL 900
901 RVPAPSVFAQ LEHIIVGEEE VMPYTFLECV NERSRILLQL VSLVTHGHER LNYWWQLLEI 960
961 PGAVQANFTR RSKRNITADA PRSHQMLQLL LAMRLTRRDI ERFPAAVHLI VAEALEEARL1020
1021 SPPMGCSMAT YELILRPELA AHAQLPFLET STGQPHCGRV YKEDSLSARC PPTGGSETDS1080
1081 PAQLRRDDMD NMDTKLLRLR FPDDMRVDEV RRLLNSSEPV VIEVQQAPGT SDHEFIEEQE1140
1141 KQLFALCSRT MTLPVGRGMF TLRTMLPRPS ESLTMPKLCL LGKEPLKGTT IEMQQIEFPA1200
1201 NMQMWPSFHN GVATGLKISP QAQDIDSNWI VYNKPKTHSH NALEHAGFLM ALGLNGHLKT1260
1261 LSFMSVYKYL VKCDEMTNVG LLLGISAAHR GTMDTKTTKL LSVHLEALLP ATAMELDIPQ1320
1321 STQVAAIMGV GLLYQGSAKR HIAEVLLQEI GRPPGPEMEN SIERESYAMT AGLSLGLVTL1380
1381 GQGESPAGLR DLQLPDTLHY YMVGGVKRPI GGSQKEKYRL ASFQVREGDT VNIDVTAPGA1440
1441 TLALGLMFFN SGNAAIAEWM QPPDSRYLLD MVRPDFLLLR TISRGLILWQ DVRPDNAWFQ1500
1501 AQFPRALRAH LKLPFYENEY APEDYDVDYE AISQAYCNIM AGAAFCIGLK YAGTENLVAF1560
1561 ATLRSVIKDF LRFPSRPMGE CAGRTTVESC LMVLLISISL VFAGSGNCEI LRIIRFLRSR1620
1621 VGPQYPHITY GSHMAIHMSL GLLFLGAGRF TISQTPESIA ALVCAFFPKF PIHSNDNRYH1680
1681 LQALRHLYVL AVEPRLFLPR DIDTNKLCLA NISVLEVGAT ELRRLPIAPC ILPVLSTLQQ1740
1741 VVVDDENYWP VCFERSRNWD QLEKALEMSA PIDIKKRTGC LSHLEDPDRL KSMLAQTLTM1800
1801 EQSICWQIDM NDLQQFASER MVKQFLSRCL DTKGTDLSPP ELMKRHQVML LFYNAVVKDR1860
1861 MHFLPVYLTL YDHVTKSMPN NIDVWQMKLI DAYLSRSQES EHPLISVELI QMMQELFKQE1920
1921 MEDSTRELCL PLREFLSRRR LDPSYVTTVS GPDLQRAFCV INYYNLLPNM LNGVDLSTGT1980
1981 VNYMRLLYEF RRLNLGAHTI FGLMKILQSL ATEVVTLDEA ALLAYTMGDQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
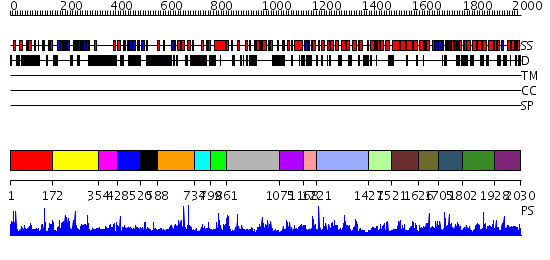
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 1.021968 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [172..353] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [354..427] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [428..519] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [520..587] | 2.033994 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [588..733] | 1.033986 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [734..798] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [799..860] | 1.043992 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [861..1074] | 1.078974 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1075..1167] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1168..1220] | 1.121988 | View MSA. No confident structure predictions are available. | |
| 12 | View Details | [1221..1426] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [1427..1520] | 2.136987 | View MSA. No confident structure predictions are available. | |
| 14 | View Details | [1521..1625] | 2.132988 | View MSA. No confident structure predictions are available. | |
| 15 | View Details | [1626..1704] | 3.133996 | View MSA. No confident structure predictions are available. | |
| 16 | View Details | [1705..1801] | 1.096996 | View MSA. No confident structure predictions are available. | |
| 17 | View Details | [1802..1927] | 1.037975 | View MSA. No confident structure predictions are available. | |
| 18 | View Details | [1928..2030] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|