













| General Information: |
|
| Name(s) found: |
CG5599-PA /
FBpp0073781
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 462 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial oxoglutarate dehydrogenase complex
[ISS]
|
| Biological Process: |
fatty-acyl-CoA biosynthetic process
[IEA]
phagocytosis, engulfment [IMP] tricarboxylic acid cycle [ISS] |
| Molecular Function: |
protein binding
[IEA]
cofactor binding [IEA] dihydrolipoyllysine-residue (2-methylpropanoyl)transferase activity [IEA] dihydrolipoamide branched chain acyltransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATHLLRNGA TCWLLRNCIS QRAALRRCLH VTSSLDKTVS FNLSDIGEGI REVTVKEWFV 60
61 KEGDTVEQFD NLCEVQSDKA SVTITSRYDG KITKIHHKID EIALVGKPLL DFDVVNEDED 120
121 EPEDSSSSSS STSSDSSASE NEEKQSAEAS ATPTEGRVII PATPSVRRLA KEHQLDLAKV 180
181 PATGKNGRVL KGDILEFLGQ VPPGTNVPHP TLLAKTPSAA PSGAASVSVP ADRVEVLKGV 240
241 RKAMLKSMTE SLKIPHFAYS DEIDMTQLMQ FRNQLQLVAK ENGVPKLTFM PFCIKAASIA 300
301 LSKYPIVNSS LDLASESLVF KGAHNISVAI DTPQGLVVPN IKNCQTKTII EIAKDLNALV 360
361 ERGRTGSLSP ADFADGTFSL SNIGVIGGTY THPCIMAPQV AIGAMGRTKA VPRFNDKDEV 420
421 VKAYVMSVSW SADHRVIDGV TMASFSNVWK QYLENPALFL LH |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
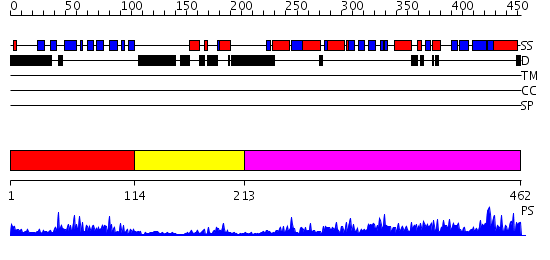
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..113] | 19.522879 | Lipoyl domain of the mitochondrial branched-chain alpha-ketoacid dehydrogenase | |
| 2 | View Details | [114..212] | 3.33 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 3 | View Details | [213..462] | 77.09691 | No description for 2ihwA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)