













| General Information: |
|
| Name(s) found: |
CG4351-PA /
FBpp0073577
[FlyBase]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 757 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Golgi cisterna membrane
[IEA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
transferase activity, transferring hexosyl groups
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGAKNRQVK NVIRPRYYSS ELGIREKLFI GVMTSQEHIN TFATAFNRTT AHLVNKIKFF 60
61 IYADSVKTNY KLKNIVGFTD TRESRRPFHV VKYIADNYLD EYDYFLLVPD TVYVDARKLV 120
121 KLLYHMSITF DLYMGGARIG LDPSGGGASA DGQSNEPPAN EEEAPGASDR NYCSLEAGIL 180
181 LSSSVIRKMR NNLERCVRIG STSDHSVNIG RCVKYASRVA GCQESFQGMR QFSYALDAPG 240
241 RRHREFSELA KEEAFRNAST VYPVQTPEDF YRLHAYYSKH HLEKVQERGY ALEQKSYRIA 300
301 NGSISNKILE IRWPLGVPPP SAPETRHDIL TWQLLNGTHN FLPNGNAEHA VATLSRIEAQ 360
361 DFAKVLEIAL QYAALKHPRL SYHSLHSAYR KFDATRGMDY QLHLNLQEGS GRSRRLVIKS 420
421 FEVVKPLGRV EVVPSPYVTE STRIAMLVPA FEHQVPDALL FVEQYERICM QNQDNTFLLL 480
481 IFMYRLESPS KGDEDPFKAL KTLALDLSSK YKTDGSRIAW VSIRLPEQLS EPVDPQSWLL 540
541 HASMYGPRQL LSLVVADLAL PKLGLESLVL LATPGMVFKA DFLNRVRMNT IQGFQVYAPI 600
601 GFQMYPCRWA HFCRECDTCD VSQSSGYFDR HNHDVIAFYS RDYVQARKLL HPQGLPIIRS 660
661 DLDIDQLLLQ PGEETRPPGV ESILDMFVAA QHSVHILRGV EPNLRFGQDV RNHLARGGTL 720
721 PQSVPERCGR EQCIHLASRK QIGDAIIRYE DKSILHK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
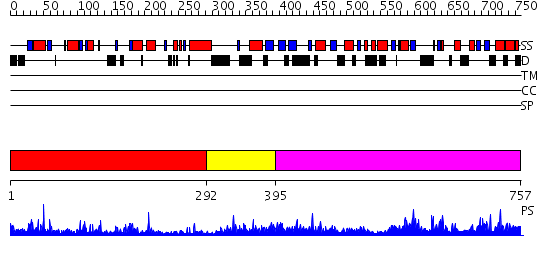
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..291] | 38.221849 | No description for 2j0aA was found. | |
| 2 | View Details | [292..394] | 1.119995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [395..757] | 1.34 | beta 1,4 galactosyltransferase (b4GalT1) |
Functions predicted (by domain):
| # | Gene Ontology predictions |
| 1 | No functions predicted. |
| 2 | No functions predicted. |
| 3 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)