













| General Information: |
|
| Name(s) found: |
CG1657-PA /
FBpp0073304
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1712 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: |
regulation of small GTPase mediated signal transduction
[IEA]
|
| Molecular Function: |
GTPase activator activity
[IEA]
Rab guanyl-nucleotide exchange factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEPPAAGNLL EIMDLARTLR QEQLFIQQEQ AAFAQLTGAF ESNAGTITKL AFVCAQQRQI 60
61 LNELLLARTD QDPLLFCRRA SAYDSAQFVD AKQLLPYEHA LAYEDLFNYL YNTPYLLALS 120
121 LATADRLSLL SASQLGQIIN TIATGLYGNA INTKDVELLL KLLRELIEIQ LLTSEQPRRL 180
181 LRTNSSSFAR LYQRLVESLF SARIFLTAAL HAPLMGVLSE HEIWLDLDPH KLMQTFTPKE 240
241 REKRFGREGD EEYQRNVARF HAETLGKLHS HVQEFVKSLQ QSWALFPSSL RWLLQTLSQQ 300
301 LRQSLRHEEQ EIRQLLTDLV FTHFISPAIA SADLLGIIDV NVSERMRHNL NQIVKLLQRL 360
361 ALNDEDSELV QLMELLMLGQ TGEDVVAILP QQSDFERSQL AINQRELAQL VEFFRLLTAR 420
421 DEYDISVEER QRLQRILGRI PKQQVQQQQQ TPKDAPDSRE SPEKSKKSNK SLMSLGKAKK 480
481 KLAKGMSFSS GSSNHNPVPV SEPLTNGQAN TSNGSGGLGA DLEHCSSHSG SNTSLSSCGA 540
541 NMPTTNDPLD MSTSSDPVLV FSLYNAGAKS KLKPLTEEEV LKMNSIGQDG SNLLPAVVTT 600
601 APLAPSSNND DVSSLEAMRR PQDDASIGNS DNLEAISEAA HSVASSLDLE EQQERDVHEN 660
661 EDNLSDMVSA NVSGRGTPNI SGRDTPSSQV TDGDGAGGDL GHHHGVHARA AANPQMQKML 720
721 LSKARSDIED KFCKFELKKF EGDENVSIIS DTWSTDVLAS DSENTLDATV SERGDRDRNF 780
781 STPLIPSAVV LPGDNNFVVE ALARAGRVSG VHGPQLDASD QRSESNWSTD VLASDSEKLA 840
841 EIDTDDNVSI TTKSDTTAPQ AAVLDDDDED EQTPGSSGDG EPDPDRGNGS ERSQEDSAFF 900
901 DAVNSYEDAN LYHGASSLAR SSVRTSYHVL GGESSFQQQY KCSGADSSGR KTTPLMGTSC 960
961 MRRQTSAESS ISNQSLNLEE PPPPMGKHHH HHREHHHRDY HHHHRERERE RERERSALKK1020
1021 KKHQQQEKEH RDLIDFSDCS EDKEELARNR DEEQPPGLVQ QLLDMINQDE QTGAVSSSSP1080
1081 NVEHRRISIE QRSAIIDGRR NGILAGSMRR HQSLNYENHE IMLNSMLPKT DDDKQEMLLC1140
1141 VQTQQLQLEE RRGSAGLRPE VDGASGIAAA GGSGLKPPTK ATGAIPKSIS FDASADKDKQ1200
1201 TYHRDGERDR ERDRERERDR ERDRERDRER DRERDHHGAG IFNKLRQGIF KNRRGASAKN1260
1261 ASNSSNQSNP AASTSSVQAD IRSVSFDPSA GCDNFGTHYC DTSEDILAKY RRKVSSSSEA1320
1321 TNSDSTGNGH GVGSTGSVGA AAHLHKHMNG GHIPGVVFGC VKQKLRTVLS RTDLHSGDFR1380
1381 QTSTTSTTMA TPLQIYLQIQ LAQCISLQRL PQISHVAEAL RCLAQLERPQ HGQLLAELQR1440
1441 DLERRQSYLQ YLMRHRQQLL LRSEQLEQLE ARLRGEARSS QRCLLQALVR MYLAWSRQQE1500
1501 KLEQFQAEFA QLRASDERVE LTEEFVESLL QELRSSADLQ DEWQVDAARV AIERMLLEQM1560
1561 YEQVMFPNED ADVSRDEVLS AHIGKLQRFV HPAHPALCIA QEYLGEAPWT FAQQQLCHMA1620
1621 AYKTPREKLQ CIINCISSIM SLLRMSSGRV PAADDLLPVL IYVVIMANPP YLLSTVEYIS1680
1681 CFLGKKLEGE DEFYWTLFGS VVKFIKTMDY LD |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
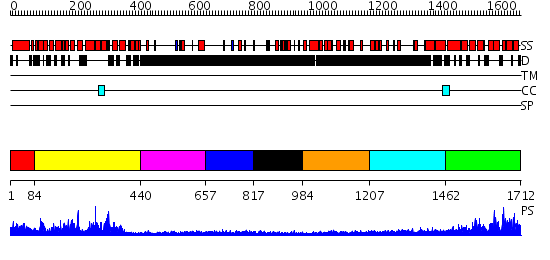
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..83] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [84..439] | 64.30103 | GAP related domain of neurofibromin | |
| 3 | View Details | [440..656] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [657..816] | 1.393996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [817..983] | 3.386993 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [984..1206] | 1.12 | Crystal structure of NimA from D. radiodurans | |
| 7 | View Details | [1207..1461] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1462..1712] | 87.0 | Crystal Structure of the Vps9 Domain of Rabex-5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)