













| General Information: |
|
| Name(s) found: |
FBpp0305231
[FlyBase]
CG32676-PA / FBpp0071432 [FlyBase] |
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1122 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cellular process
[IMP]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKSAATNSA TADGQNEAAA AAAAAATAAG CGSNNSSSTS SSNNTANSNN ALQRLATTTS 60
61 AVVASPMTIN LNISTTTGGN FGVSVEPHIS VESLKKIIAK KLKVAKDRIC LLHREKELQD 120
121 GTLRDHNLMD GSKIILIPNV ETGLLAQRPE NTVMQALESL NDAQVNDFLS GKTPLNLSMR 180
181 LGDHMMLIQL QLSTVNPAGG GAATVAPVAG GASGSSINAA ASTTSTVPVM PAGTGCSGAN 240
241 STSLSSSAVL AAAAAGVGGS TGTSGSGTSS SSSSTSSSSS SSSSSSRTRS SGQRSSGRIG 300
301 HGHVHSHQHP SLHAANWHGF SHGHGHGHSH GNGHGHHHHH HHHHHHHHHH HNASAVAAGS 360
361 GGGMSSLRKM YGDLPSSSGA SGSAPATGTG QSQSSSTLNG PDLTKLLIKG GIQNIVNSFA 420
421 LKDAGANARP VANGGSGSSP GKPVLEQSPI KSLSNLVSSP IKMTTIKNIP LPTTAATSSS 480
481 SSCNCSSAAP SSSSSTSSGA CATGGCQQDP IAAKLTSCLC TRLPTPSVAP PVATLVAPAP 540
541 VARSGSTSSP SKCPESSCSG ANAGFAARST LAGTRVTFSG NSMLHKTGNN RITRTKHRHY 600
601 HGQGHGHGHG NGHSSSSSSS SSSSSSHFFN HACAGNQPSV NAAAFATSSS SSSSSPSSSS 660
661 SSPSKRRKLE QGMGAGAVGA TAATATAAAT VALPASTSAT TAEPDDSLLG VSDTRTLAEA 720
721 SRNLTQTLRK LSKEVFTNKI DLSGAGVVSS GRSSSSGATS SGAASGEEAP RKSGSGAVIE 780
781 SMKNHGKGIY SGTFSGTLNP ALQDRFGRPK RDISTVIHIL NDLLSATPQY GRGARISFEA 840
841 PTTGSVSGSG SGSGSSGSSS TTSSGSGLHH GSRSKMYSSK HHNCAKCNSR AQQQQQQSAT 900
901 LASGSGGSCS FRDCPSYHSS SAATKASTSS KSSSHSCCQT AEAPSSMTSQ SNGCTCRYRR 960
961 DDGQGERERE HTCQKCTVEL ANMKTRSKMD QLRLVMQQHK QKREARKLKS GPYATAASAS1020
1021 AADSLSSIAA GGTAAVPMGA AAQYSAVSAL VATPLQPAAV PVQTGKAAPA NSITGNSSNA1080
1081 NVNGNTSTAP ATAATSAAAA PTAAPPSEVS PNHIVEEVDT AA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
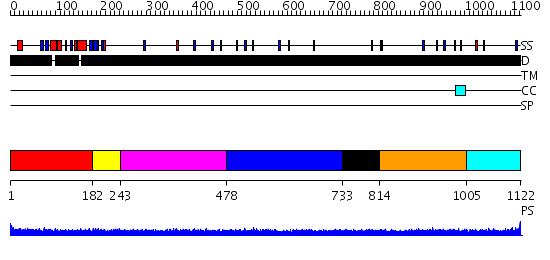
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..181] | 2.69 | Crystal Structure of ISG15, the Interferon-Induced Ubiquitin Cross Reactive Protein | |
| 2 | View Details | [182..242] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [243..477] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [478..732] | 1.124997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [733..813] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [814..1004] | 2.115983 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1005..1122] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. | |||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.28 |
Source: Reynolds et al. (2008)