













| General Information: |
|
| Name(s) found: |
FBpp0309355
[FlyBase]
Tbh-PB / FBpp0089042 [FlyBase] |
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 670 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
regulation of behavior
[IMP]
histidine catabolic process [IEA] ovulation [IMP] male courtship behavior [IMP] larval locomotory behavior [IMP] oxidation reduction [IEA] regulation of forward locomotion [IDA] behavioral response to ethanol [IMP][TAS] catecholamine metabolic process [IEA] octopamine biosynthetic process [TAS] memory [IMP] |
| Molecular Function: |
copper ion binding
[IEA]
dopamine beta-monooxygenase activity [IEA][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLKIPLQLSS QDGIWPARFA RRLHHHHQLA YHHHKQEQQQ QQQQQQQQQA KQKQKQNGVQ 60
61 QGRSPTFMPV MLLLLMATLL TRPLSAFSNR LSDTKLHEIY LDDKEIKLSW MVDWYKQEVL 120
121 FHLQNAFNEQ HRWFYLGFSK RGGLADADIC FFENQNGFFN AVTDTYTSPD GQWVRRDYQQ 180
181 DCEVFKMDEF TLAFRRKFDT CDPLDLRLHE GTMYVVWARG ETELALEDHQ FALPNVTAPH 240
241 EAGVKMLQLL RADKILIPET ELDHMEITLQ EAPIPSQETT YWCHVQRLEG NLRRRHHIVQ 300
301 FEPLIRTPGI VHHMEVFHCE AGEHEEIPLY NGDCEQLPPR AKICSKVMVL WAMGAGTFTY 360
361 PPEAGLPIGG PGFNPYVRLE VHFNNPEKQS GLVDNSGFRI KMSKTLRQYD AAVMELGLEY 420
421 TDKMAIPPGQ TAFPLSGYCV ADCTRAALPA TGIIIFGSQL HTHLRGVRVL TRHFRGEQEL 480
481 REVNRDDYYS NHFQEMRTLH YKPRVLPGDA LVTTCYYNTK DDKTAALGGF SISDEMCVNY 540
541 IHYYPATKLE VCKSSVSEET LENYFIYMKR TEHQHGVHLN GARSSNYRSI EWTQPRIDQL 600
601 YTMYMQEPLS MQCNRSDGTR FEGRSSWEGV AATPVQIRIP IHRKLCPNYN PLWLKPLEKG 660
661 DCDLLGECIY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
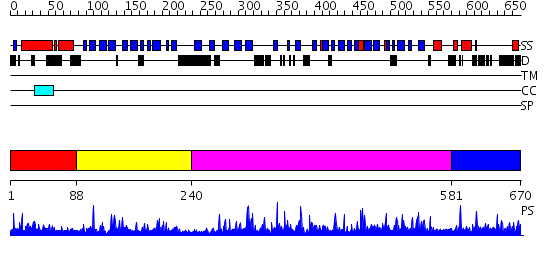
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [88..239] | 1.69 | Cytochrome domain of cellobiose dehydrogenase | |
| 3 | View Details | [240..580] | 97.69897 | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase with bound peptide and dioxygen | |
| 4 | View Details | [581..670] | 1.061993 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|