













| General Information: |
|
| Name(s) found: |
CG5937-PA /
FBpp0070833
[FlyBase]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2016 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: |
regulation of ARF protein signal transduction
[IEA]
|
| Molecular Function: |
ARF guanyl-nucleotide exchange factor activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALDTKRPKF ITMGLNGLHR VIKDERFYIG LEPEDDSVWL PSQLLRATNG ILPTTSSEDT 60
61 VVNVLRLFLA MACSPACTLN GRLLIEILSR CGECWEMGSR ATKAASLAAA SQCLRTFCAF 120
121 LIEEAEEVKK TAPAGLMTQT QASAVYNEVI PVMQWLCSRL VEPNVNASPN KKCENHSSLY 180
181 LTECILTLSS ALPRNVHANP HFTSFLWQKF CPTLAAALGS PGRINLDKKF TYKLVKGNKN 240
241 KNIESPNIHY LRDALHMIEN EARGFFTGPG LDGPQARCVY LTAIQLLRIA GAHGSLRPML 300
301 EALFHRMLLL PAPQNRTEPL RCVREIFKSP ERLIDLAVIL YVDKNTAQGC SDEMALFRLL 360
361 VDAMEECAYG ASGGGSATEA SLQASVECMV ALLDSLQVLC SGELTESMIS DQIVQVVNGR 420
421 HDLLKDADYS GPLTYQSMAR LPAPYRDAIV EFRQTVFETS SGSESEGEPQ HEQDAASNGS 480
481 GDTEGPEDDD PSSSSDETSR VENRWPYSHL EAPVMPIRTD SDNDRQHARD FARALRQDLV 540
541 PKLLRLRSCV ELDEAMQEFA SAVCQENSMN FSDFDYNLTA INADGIYLAI YSSLLLSLQL 600
601 MRAGFYEQVA QGMSHKDILV PMSEQQFVTS VQNTGVLVYL SSPWLCELYQ SVTVCNVLEA 660
661 MPRQQLEGMG PRCALVDMLC DAGGLGATQM LSEWQRLQTA NVKHKEEEQL HDKRREAAKK 720
721 LCRRLLTCCW DSMVIVLSSG LGDLQTSSAS NKLVALSKRT LRVKAKANKS NGEALYAMCL 780
781 DGLHSAATLS NSLNLQHLAG KILNLLASNV CQTSGPRISA SQAMSMDVVL TGGLNLGSYS 840
841 ADCWPSIFAV CRHVSQLEHE IFSMQNPAIS ASPGSSRRDL ETGEKLSNGN AQDKLNLSSI 900
901 PIDDDETCVD VYSFLQAPMQ SPNTNITSIL KVYSGTNETV LLSQSDTSKV LCALSHQAEN 960
961 LFGDAAERLS LPSLCQFLKH LCRASRDQLY KSQVARKGSR IWWPSKGWKK LDSLPMSLLL1020
1021 HRIGDVTLKV FRSSRPLLHV LKVWAITGPH LMDAACHRDR MISKRAIEYI HDIITALLVE1080
1081 QSELPYFHFN EALLKPFENL LSMDTDVDVQ DQIVACLYEV VEAHRTEIRS GWRPLFGTLR1140
1141 NARSRMLNMS NIIDIFRVFL DSDNTLVFAN AGLDCILCLL SYLEISGGGN NNACSGGSGS1200
1201 AAGGTASNGG QQEEDNTFRP TDFLHETLRF LERCSSILGF MHSMPKCPNF HSTYKIKGIS1260
1261 YTHIIDANIP SSMENFTYFG NDYLQTRNEQ YMISYRSLHI DKDTIVKIDE MDKPSGVLKV1320
1321 WFLLLDGLTN SLIVCPYSHQ APILQTIFKL FKNLLASPGI DFGFYCINHL LVPMIQDWLR1380
1381 YINKTGSSWQ LVEKNFKHCC CMTTDLVVEF IEKSVPEQRR LGAGTKTRLA QIVHPADNLL1440
1441 YSKLKFVTER IEREQQQQQS PSVQDHGCGY TQQYDSSSSS ASEEQQKNGG GVGGGGSNLI1500
1501 ESPTKISGSA TLALKQLLLV LIECAAQSQE AIARISVSCL KHVILSTGML FNESQWMIAC1560
1561 SAIHRACTVT IAPLRQLSFA FHEKSNSFYG DCANVKVAAR RDSSLEELAR IYALAQQVFL1620
1621 SDNQREPGQN QAPTPSASQC KLSDDRSYSF LLYPLNNGFN SNLDNFVIRI PFKNLVVGLL1680
1681 ANQMLLQLVA KLLLSRLKCV PQAVSTCIFD NYAASAASPS HDYDLDFRSK EILLRCVKQY1740
1741 LMSALEFDSR PGLKFLMQKV SNIEYAANLY KQMTSSWMIY YIALVDSHLN DIVVYNLGPE1800
1801 DLNFILESCS RLNTTTVKKK ENFVRYLFCL QDAWNLVCEL YLSNSALHEI ESGSGKLRQK1860
1861 PPQLLHHPQT KPMCISLNGN GDAEMTDTGS GSGSGSSTSP SKCVQLQEEE NVTMTTLISE1920
1921 FQPKCRSNPF DTNRQAKSSE AESISPEIEQ QRASSILKDS NYKRAALAQL VVASMELLRS1980
1981 LPGEAEENLK LLMTPTIREA FRLVQLQGNE LKVNQF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
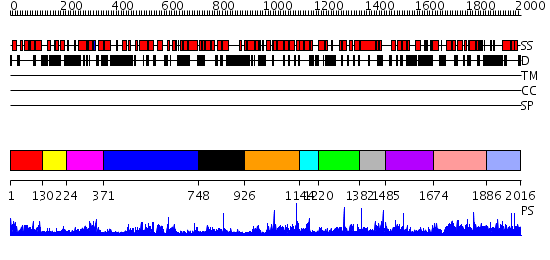
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..129] | 6.124985 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [130..223] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [224..370] | 1.158975 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [371..747] | 42.39794 | The structure of RalF | |
| 5 | View Details | [748..925] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [926..1143] | 2.142972 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1144..1219] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1220..1381] | 2.110985 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1382..1484] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1485..1673] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1674..1885] | 2.028988 | View MSA. No confident structure predictions are available. | |
| 12 | View Details | [1886..2016] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 |
|
|||||||||||||||
| 5 | No functions predicted. | |||||||||||||||
| 6 | No functions predicted. | |||||||||||||||
| 7 | No functions predicted. | |||||||||||||||
| 8 | No functions predicted. | |||||||||||||||
| 9 | No functions predicted. | |||||||||||||||
| 10 | No functions predicted. | |||||||||||||||
| 11 | No functions predicted. | |||||||||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)