













| General Information: |
|
| Name(s) found: |
CG4199-PA /
FBpp0070323
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 552 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
|
| Biological Process: |
cell redox homeostasis
[IEA]
oxidation reduction [IEA] |
| Molecular Function: |
FAD binding
[IEA]
oxidoreductase activity [IEA] 2 iron, 2 sulfur cluster binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTEESSPDS EYTSAVPVDC RVTDLKENEM KQVDFDEDTR VLLVKQNDRL LAVGAKCTHY 60
61 GAPLQTGALG LGRVRCPWHG ACFNLENGDI EDFPGLDSLP CYRVEVGNEG QVMLRAKRSD 120
121 LVNNKRLKNM VRRKPDDQRV FIVVGGGPSG AVAVETIRQE GFTGRLIFVC REDYLPYDRV 180
181 KISKAMNLEI EQLRFRDEEF YKEYDIELWQ GVAAEKLDTA QKELHCSNGY VVKYDKIYLA 240
241 TGCSAFRPPI PGVNLENVRT VRELADTKAI LASITPESRV VCLGSSFIAL EAAAGLVSKV 300
301 QSVTVVGREN VPLKAAFGAE IGQRVLQLFE DNKVVMRMES GIAEIVGNED GKVSEVVLVD 360
361 DTRLPCDLLI LGTGSKLNTQ FLAKSGVKVN RNGSVDVTDF LESNVPDVYV GGDIANAHIH 420
421 GLAHDRVNIG HYQLAQYHGR VAAINMCGGV KKLEAVPFFF TLIFGKGIRY AGHGSYKDVI 480
481 IDGSMEDFKF VAYFINEADT VTAVASCGRD PIVAQFAELI SQGKCLGRGQ IEDPATREDW 540
541 TKKLGQPLPQ VR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
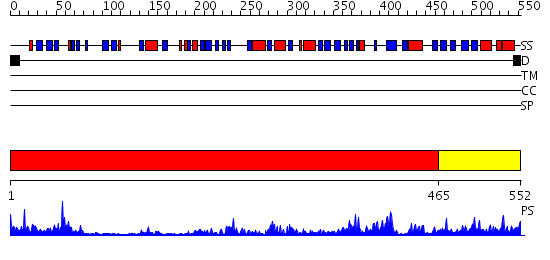
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..464] | 22.221849 | Dihydropyrimidine dehydrogenase, N-terminal domain; Dihydropyrimidine dehydrogenase, domain 4; Dihydropyrimidine dehydrogenase, domain 3; Dihydropyrimidine dehydrogenase, domain 2; Dihydropyrimidine dehydrogenase, C-terminal domain | |
| 2 | View Details | [465..552] | 64.0 | Crystal Structure of Putidaredoxin Reductase from Pseudomonas putida |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)