













| General Information: |
|
| Name(s) found: |
gi|24639248
[NCBI NR]
gi|22831547 [NCBI NR] |
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 647 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
|
| Biological Process: |
cell redox homeostasis
[IEA]
oxidation reduction [IEA] |
| Molecular Function: |
FAD binding
[IEA]
oxidoreductase activity [IEA] 2 iron, 2 sulfur cluster binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVWQSPRTA LSLVKTKRAF EGLLRIRKLP SSQSWRILNA PSFASHINNR LLNTMGSVNC 60
61 KEYKTTGSTP KKEKSTGGAV GSYQSKCSNQ SPSTTMSTEE SSPDSEYTSA VPVDCRVTDL 120
121 KENEMKQVDF DEDTRVLLVK QNDRLLAVGA KCTHYGAPLQ TGALGLGRVR CPWHGACFNL 180
181 ENGDIEDFPG LDSLPCYRVE VGNEGQVMLR AKRSDLVNNK RLKNMVRRKP DDQRVFIVVG 240
241 GGPSGAVAVE TIRQEGFTGR LIFVCREDYL PYDRVKISKA MNLEIEQLRF RDEEFYKEYD 300
301 IELWQGVAAE KLDTAQKELH CSNGYVVKYD KIYLATGCSA FRPPIPGVNL ENVRTVRELA 360
361 DTKAILASIT PESRVVCLGS SFIALEAAAG LVSKVQSVTV VGRENVPLKA AFGAEIGQRV 420
421 LQLFEDNKVV MRMESGIAEI VGNEDGKVSE VVLVDDTRLP CDLLILGTGS KLNTQFLAKS 480
481 GVKVNRNGSV DVTDFLESNV PDVYVGGDIA NAHIHGLAHD RVNIGHYQLA QYHGRVAAIN 540
541 MCGGVKKLEA VPFFFTLIFG KGIRYAGHGS YKDVIIDGSM EDFKFVAYFI NEADTVTAVA 600
601 SCGRDPIVAQ FAELISQGKC LGRGQIEDPA TREDWTKKLG QPLPQVR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
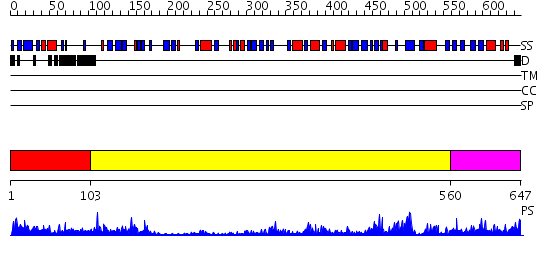
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | 1.143995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [103..559] | 18.0 | Dihydropyrimidine dehydrogenase, N-terminal domain; Dihydropyrimidine dehydrogenase, domain 4; Dihydropyrimidine dehydrogenase, domain 3; Dihydropyrimidine dehydrogenase, domain 2; Dihydropyrimidine dehydrogenase, C-terminal domain | |
| 3 | View Details | [560..647] | 63.39794 | Crystal Structure of Putidaredoxin Reductase from Pseudomonas putida |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)