













| General Information: |
|
| Name(s) found: |
FBpp0292166
[FlyBase]
FBpp0292165 [FlyBase] east-PB / FBpp0070312 [FlyBase] |
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2301 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][NAS]
[NOT]nuclear chromosome [IDA] [NOT]nucleolus [IDA] |
| Biological Process: |
mitotic metaphase
[IMP]
achiasmate meiosis I [IMP] proteolysis [IEA] female meiosis chromosome segregation [IMP] |
| Molecular Function: |
zinc ion binding
[IEA]
metallocarboxypeptidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSRKVPGGS GGADESTAAA APLDDNANAS VEIPDSSEEP AMGVGEEMSI ISKTRTSTLS 60
61 VEPAKEPTVT AELEGEKELE SNPVSKTPRS TPTPTLTPAV TPTASDGVAA KSVRVTRHSS 120
121 PLLLIISPTT SRREVGDGEL DTEEPTGSGG QRKSSVERSL APVIRGRKSI KDLKEAKEVK 180
181 SEEPPAAASE SRAASGVTPG QVKEQHVADG NEMESLPITD KKDHKDTKDK GDERETDQEE 240
241 EKEKSADTEI IADTEKTSEK QKYTEKDKAA DKDGGKEKDI DANKDIDKEK EKVKEVLPPV 300
301 VPIAPVTPTC NRVTRKSHAQ EQAINTRVTR NRRQSSTVGA NSTASLVAAS SSVTEQPPPS 360
361 RGRRKKPVVV APPLEPAVKR KRSQDVEADS DANNSTKYSK VEVVKSEEAE APEEDSSAVP 420
421 IKQESVDGNE VSSISPTVTP TPTPAPTPAP VPGSRRGRGR PQNRNSSSPA TTTRATRLSK 480
481 AGSPVILTPV AQEPAPPKRR RVGSSTRKTV SASSLAPSSQ GGAGDEDSKD SMASSMDDLL 540
541 MAAADIKQEK LTPDFDDSLM PEGLPSTSGA SSANGHSCTE PLTVDTEINV KPADSKVKPK 600
601 ESPVVAVEES PSQSETQSAK VSAHAGKAPS LSPDMISEGV SAVSVRKFYK KPEFLENNLG 660
661 IEKDPELGEI VQTVSNNDTE TDVEMAVDGE VNQPSTPKSQ DKKKEEQEKN QKSGLKAAKK 720
721 APAKLEPKAE DISEILTDVP VDISTEAVEI IEEAEEDTCS NSSIKPGELR LDESNDEPEL 780
781 LLEDALIVNG DENETPDQPE EKEDQVEFFH TGEYDDFEHE IMVELAKEGV LDASGNALSQ 840
841 QKVELEHPED VTLHESKNDI EAEESVERKP LKDPSVADEM EDMNEESYID IKDQTNQLLV 900
901 EHLAEEAMEA DCGPEDNKEN LSTSASSTAA DGLDIQLAIK EDDDEEKPLA VIADEQKPGL 960
961 LLTNDMKVDE KPNGKQESVC DEHVQLVPNL RQEQEIHLQN LGLLTHQAAE HRRKCLLEAQ1020
1021 ARQAQMQLQQ HHHHQHKRQG ARGGGSATHV ESSGTLKTVI KLNRSSNGGV SGSGGLPTGT1080
1081 VIHGGCGSSS ASSTSSSSVG SATRKSSGTL GSGAGAGAGV RRQSLKMTFQ KGRARGHGAA1140
1141 DRSADQYGAH AEDSYYTIQN ENEDSSHSES QGQTDHGFYQ MVKKDEKEKI LIPEKASSFK1200
1201 FHPGRLCEDQ CYYCSGKFGL YDTPCHVGQI KSVERQQKIL ANEEKLTVDN CLCDACFRHV1260
1261 DRRANVPSYK KRLSASGHLE MGSAAGSALE KQFAGDSGVI TESGGEAGST AAVAVQQRSC1320
1321 GVKDCVEAAR HSLRRKCIRK RVKKYQLSLE IPAGSSNVGL CEAHYNTVIQ FSGCVLCKRR1380
1381 LGKNHMYNIT TQDTIRLEKA LSEMGIPVQL GMGTAVCKLC RYFANLLIKP PDSTKAQKAE1440
1441 FVKNYRKRLL KVHNLQDGSH ELSEADEEEA PNATETERPT SDGHEDPEMP MVADYDGPTD1500
1501 SNSSSSSTAA LDTSKQMSKL QAILQQNVGA DAAGAAGTGT VAASPGGSGS GADISNVLRG1560
1561 NPNISMRELF HGEEELGVQF KVPFGCSSSQ RTPEGWTRVQ TFLQYDEPTR RLWEELQKPY1620
1621 GNQSSFLRHL ILLEKYYRNG DLVLAPHASS NATVYTETVR QRLNSFDHGH CGGLNIAGSP1680
1681 SSSGSGKRSG VPQPTGASVL ATALTTPLTS HSSSSASISS EQHSSVDPVI PLVDLNDDDE1740
1741 GEDGAGGAGE RESTNRQQDV ILECLRTASV DKLTKQLSSN AVTIIARPKD KSQLSCNSGS1800
1801 STSISSSSSA ISSPEEVAVT KVTAVAPVQS KDAPPLAPAS SGVSNSRSIL KTNLLGMNKA1860
1861 VELVPLTTAP HAYKPTGCHK PEKQQKILDV ANKQPGSQGE PVPSSALLGL QSKLKPPTHQ1920
1921 QQVSGSGAGT SGSQKPSNVA QLLSSPPELI SLHRRQTSGA AAGSSSFLQG KRLQLPRSGA1980
1981 GPSGAGTGTG AGAAGSRSAG GPPPPNVVIL PDALTPQERH ESKSWKPTLI PLEDQHKVPN2040
2041 KSHALYQTAD GRRLPALVQV QSGGKPYLIS IFDYNRMCIL RREKLMRDQL LKSNAKPKPQ2100
2101 NQQQQQGQTH QQQQNSAASA AAFSNMVKLA QQHTARQQLQ QLQQKPQQQQ QLPTLQPGGV2160
2161 RLARLPQKLL MPPLTNPQIG SQAPNLQPLL SSTLDNSNNC WLWKNFPDPN QYLLNGNGGG2220
2221 AGSSSSKLPH LTAKPATATS SSGAANKSAG SLFTLKQQQH QQKLIDNAIM SKIPKSLTVI2280
2281 PQQMGGNTGG DMGGSSSSGK D |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
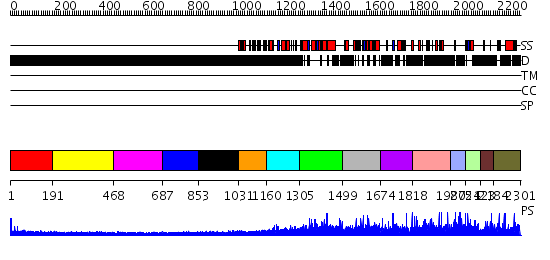
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | 2.335997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [191..467] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [468..686] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [687..852] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [853..1030] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1031..1159] | 0.982 | Thermus thermophilus Leucyl-tRNA synthetase complexed with with a tRNAleu transcript in the post-editing conformation and a post- transfer editing subsrate analogue | |
| 7 | View Details | [1160..1304] | 2.292996 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1305..1498] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1499..1673] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1674..1817] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1818..1986] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1987..2053] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [2054..2122] | N/A | No confident structure predictions are available. | |
| 14 | View Details | [2123..2183] | N/A | No confident structure predictions are available. | |
| 15 | View Details | [2184..2301] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)