













| General Information: |
|
| Name(s) found: |
trr-PD /
FBpp0070347
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2431 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][NAS]
polytene chromosome [IDA] |
| Biological Process: |
histone methylation
[IDA]
smoothened signaling pathway [NAS] positive regulation of transcription [IGI] progression of morphogenetic furrow during compound eye morphogenesis [IGI][IMP] compound eye development [IMP][NAS] |
| Molecular Function: |
histone methyltransferase activity (H3-K4 specific)
[IDA]
DNA binding [IEA][NAS] zinc ion binding [IEA] transcription coactivator activity [IGI] histone methyltransferase activity [NAS] receptor binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNIPKVTTSL GAAEKAKPER VASVAAAAFN AVSLQKRSGD DTATPAEDPT RKKAKTELLL 60
61 GTGTAAPSLP AKASSTAPQQ LLYQRSGQQA KAQVKAASEP QDVETADGVW DARDQQIIVC 120
121 NFGSGTEMGA IKAEDADKQS EYRISTPRNS QSNPLLHRNT AFTSFTKKEG ASSSASSSSS 180
181 TASVISIEPS GSGQDHAENS GKSEDLDYVL MPASGADSST SVGNSTGTGT PAGTPIGATT 240
241 STIILNANNG TAGVSGAGTT TILTQKSGHT NYNIFNTTAT GSQTPTTTLL NRVNLHPKMK 300
301 TQLMVNAKKL SEVTQTTAKV SIGNKTISVP LLKPLMSASG AATAGGATIV ESKQLLQPGG 360
361 QVTTVMSAAQ QSGGQQVHPH VHSHAHHNFT KLIKRGPKNS GTIVSFSGLQ IKPANTKIVA 420
421 TKVVSKKMLQ LQQHQQQIQQ QQQLQQLQVT SGGGLAPPTG SIVTITTTNP SQTYAMVQDS 480
481 ATVGPAAHSE DDAPAPRKIT AYSENLQKIL NKSKSQESTG GPEEFTNINS VVIKPLDKNT 540
541 LNCPPSFNIF KQQQHSQAAQ SQSISAVGSG AGTPVTFTMA SGNASDLATT STVSVSAGTI 600
601 CINSPMMGTR PIISIQNKNI SLVLSKTTMA QQKPKMITTT TLSSQAALQM HHALIQDSSA 660
661 DKAGSSANSG SATSGASMQL KLTTANTPTK LSVSLAPDVV KLEEVGSESK AKLLVKQEAV 720
721 VKDSTGTPTS EERAEEIGTP EKRLNANATM TAINQVQNQS ANQIQMATST STASNPSTPN 780
781 PTVNATPMNN QRSAAEDNAL LKQLLQNNSS SHSLNQISIT SAHVGSASAS APLSARKVIN 840
841 VRAPSMGKVR SLEDQLARPV IPPVPTATQA AGSSSSSGSV ATSTTTTTVA SGGSSQQVAT 900
901 ASATALPVSA VAITTPGVGG EAKLEQKSDQ PAAIMQNQSQ NQAPPPPPPP QQQQQQQLHQ 960
961 PQQLQPSPHQ VKQTVQIVSK ETSFISGPVA AKTLVTEATS KPAELLPPPP YEMATAPISN1020
1021 VTISISTKQA APKELQMKPK AVAMSLPMEQ GDESLPEQAE PPLHSEQGAT AAGVAPHSGG1080
1081 PLVSAQWTNN HLEGGVATTK IPFKPGEPQK RKLPMHPQLD EKQIQQQAEI PISTSLPTTP1140
1141 TGQGTPDKVQ LISAIATYVK KSGVPNEAQP IQNQSQGQVQ MQAQMQATMQ GHLSGQMSGQ1200
1201 ISGHAAGQIP AQMHLQVQHQ LHMAVHPQQQ QQQLHQNQPQ NATIPLPVTG QGAVPIPVPT1260
1261 MESKAGDQRK RRKREVQKPR RTNLNAGQAG GALKDLTGPL PAGAMVQLAG MPPGTQYIQG1320
1321 AASGTGHVIT STGQGVTLGG VGASTGASSS PMLKKRVRKF SKVEEDHDAF TEKLLTHIRQ1380
1381 MQPLQVLEPH LNRNFHFLIG SNETSGGGSP ASMSSAASAG SSSAGGGKLK GGSRGWPLSR1440
1441 HLEGLEDCDG TVLGRYGRVN LPGIPSLYDS ERFGGSRGLV GGSARTRSPS PAESPGAEKM1500
1501 LPMSSIQNDF YDQEFSTHME RNPRERLVRH IGAVKDCNLE TVDLVESEGV AAWATLPRLT1560
1561 RYPGLILLNG NSRCHGRMSP VALPEDPLTM RFPVSPLLRS CGEELRKTQQ MELGMGPLGN1620
1621 NNNNNYQQKN QNVILALPAS ASENIAGVLR DLANLLHLAP ALTCKIIEDK IGNKLEDQFM1680
1681 NQDDEKHVDF KRPLSQVSHG HLRKILNGRR KLCRSCGNVV HATGLRVPRH SVPALEEQLP1740
1741 RLAQLMDMLP RKSVPPPFVY FCDRACFARF KWNGKDGQAE AASLLLQPAG GSAVKSSNGD1800
1801 SPGSFCASST APAEMVVKQE PEDEDEKTPS VPGNPTNIPA QRKCIVKCFS ADCFTTDSAP1860
1861 SGLELDGTAG AGTGAGPVNN TVWETETSGL QLEDTRQCVF CNQRGDGQAD GPSRLLNFDV1920
1921 DKWVHLNCAL WSNGVYETVS GALMNFQTAL QAGLSQACSA CHQPGATIKC FKSRCNSLYH1980
1981 LPCAIREECV FYKNKSVHCS VHGHAHAGIT MGAGAGATTG AGLGGSVADN ELSSLVVHRR2040
2041 VFVDRDENRQ VATVMHYSEL SNLLRVGNMT FLNVGQLLPH QLEAFHTPHY IYPIGYKVSR2100
2101 YYWCVRRPNR RCRYICSIAE AGCKPEFRIQ VQDAGDKEPE REFRGSSPSA VWQQILQPIT2160
2161 RLRKVHKWLQ LFPQHISGED LFGLTEPAIV RILESLPGIE TLTDYRFKYG RNPLLEFPLA2220
2221 INPSGAARTE PKQRQLLVWR KPHTQRTAGS CSTQRMANSA AIAGEVACPY SKQFVHSKSS2280
2281 QYKKMKQEWR NNVYLARSKI QGLGLYAARD IEKHTMIIEY IGEVIRTEVS EIREKQYESK2340
2341 NRGIYMFRLD EDRVVDATLS GGLARYINHS CNPNCVTEIV EVDRDVRIII FAKRKIYRGE2400
2401 ELSYDYKFDI EDESHKIPCA CGAPNCRKWM N |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
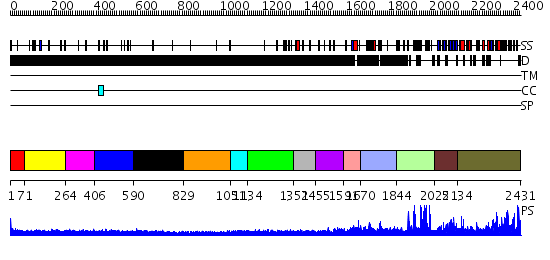
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [71..263] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [264..405] | 1.160987 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [406..589] | 1.158986 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [590..828] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [829..1050] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1051..1133] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1134..1351] | 1.136988 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1352..1454] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1455..1590] | 1.101987 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1591..1669] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1670..1843] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [1844..2024] | 1.14 | No description for 2jm1A was found. | |
| 14 | View Details | [2025..2133] | N/A | No confident structure predictions are available. | |
| 15 | View Details | [2134..2431] | 66.09691 | No description for 2r3aA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)