













| General Information: |
|
| Name(s) found: |
FBpp0311616
[FlyBase]
A3-3-PA / FBpp0070234 [FlyBase] |
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 613 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
nervous system development
[IMP]
regulation of transcription, DNA-dependent [IEA] |
| Molecular Function: |
transcription factor activity
[IEA]
protein dimerization activity [IEA][ISS] sequence-specific DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFNSNIPASS LLSIDSGGLL MGGHTPKTPE ILNSLIAMTN PLENFSYSSA AAAAAAVSVA 60
61 SSSASCSAAS TPVVTVRHFN GHPNGQSHSQ DSSHSSCSGS PLDSPAGTAT TPSVQQTCSR 120
121 LIKEGLKLSI QSKRKLSTCD SSSGSEQPHS KYSRRSSNHN GHSGSSNNYS GSMSNANDLD 180
181 DDCEESSDDD SETKSQPKGL TPEDEDRRRR RRERNKIAAT KCRMKKRERT QNLIKESEVL 240
241 DTQNVELKNQ VRQLETERQK LVDMLKSHGC QRAGGCQLPS QLLQSPAQKY LSELELETVS 300
301 IDGPNSGNNN QRLQSIPSMA TFKYGSKTAA AMAQQLPNGY CKPSPSAQEF EHAGYQQQQQ 360
361 QQQQQQPQSL NPAGNNVIDQ QHANPSPSLL SDYVPNCDGL TGSASNHPSH NNNNNNNNSS 420
421 GASSNTSNNN SNISSHSSNA TSSTTPTATS SAIEFVKNEL VDSQSPYTTA LSAERFLFEP 480
481 SDGFPDIKHA CVLPSPCGIN GLNMASVVNT NGSTGNNNNS HHNNNNNNNN HLMDFHQGLQ 540
541 HGNIGTGVGV GVGVGVGVGV GVGVMTPYDD EQLLLLKNGC FATDLLSQLV EEGGEYVDLD 600
601 SATAFMNNGS CLA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
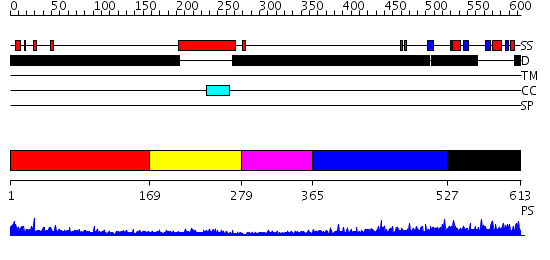
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..168] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [169..278] | 8.221849 | Crystal Structure of Human NFAT1 and Fos-Jun on the IL-2 ARRE1 Site | |
| 3 | View Details | [279..364] | 1.02 | MJ0307, thioredoxin/glutaredoxin-like protein | |
| 4 | View Details | [365..526] | 3.180997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [527..613] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.46 |
Source: Reynolds et al. (2008)