













| General Information: |
|
| Name(s) found: |
Slip1-PA /
FBpp0088159
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 767 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
protein binding
[IPI]
potassium channel regulator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIADVEYEY VVLKINGYDI SHLSRYEAVQ KFLQSKETLV VEIRRQKHNA LDLELKHGSN 60
61 AKISKVDNPG ELSVLTDKSA EGTITAASAS QQINCPSSTS LKEIETKTPV VLTLRARSHE 120
121 DRLGSLQAAS KETQTQSVVG TDVLKDNDLV NTITDNFIEH EHHLFEQCLE PEIDIEEVTL 180
181 VKGVEQSSSN QIGLIVTSSG IQQSSTDTNK GDILGNVLEH SEDVFISGVQ PESIAYRDGR 240
241 LRQGDQILRI NGLDVKNQEE LETQIARSST SVTLLVSRIL YPEDDDDEDI HFEYANTFLP 300
301 DDYTNVVDKL DKVLLTHVKS LEELSNKSAM QSDECYHIPE KNSSDSNVKI SSLAKNIIEQ 360
361 STKSCSKIKL RPNANLDYKK KFNLPLSQEE VHLQYEYDES EHIYETIPED SESEPVYCSP 420
421 YQRSNDKTSI GCSSPIASRP AESLERTMQQ QTQRVAQWLG LKPQYQKTRQ TLVGRPPPLK 480
481 LVQQPTCSRV FTLRSTLTNT SASSSSGVAY SSYGQNNVVT GNAAAPGEEV DNSSSAYNTG 540
541 DSNNSASPHQ NTTNPDEAIA TGRKLDSTVI DSPNDHLDAT GVSTMLLLPF GKSGRIGLCS 600
601 SNLPTAYVSE RYTNVGSENE IHPLKSDIEI LRVKPTDDSY SHCPQFNAPN LSSYHFVSSQ 660
661 EVANRCHIST SLQKNATLLN GESAEEIPMV WKVKRRPDGT RYIVKRPVRN RPQVALRKNM 720
721 RYNEVTTTED DTISEVKIGR YWTKEERKRH IERAREKRHH QTQQQQQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
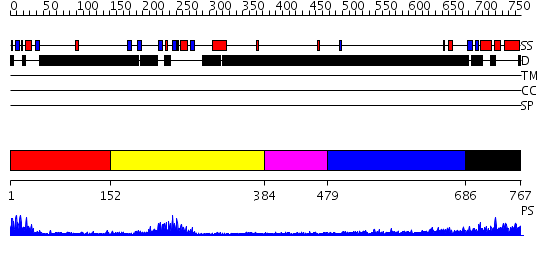
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | 8.69897 | Structural Insights into the Inter-domain Chaperoning of Tandem PDZ Domains in Glutamate Receptor Interacting Proteins | |
| 2 | View Details | [152..383] | 17.221849 | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | |
| 3 | View Details | [384..478] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [479..685] | 1.294997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [686..767] | 2.175995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)