













| General Information: |
|
| Name(s) found: |
FBpp0306758
[FlyBase]
Axn-PC / FBpp0084920 [FlyBase] |
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 739 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGHPSGIRK HDDNECSGPR PPVPGEESRV KKMTEGVADT SKNSSPSYLN WARTLNHLLE 60
61 DRDGVELFKK YVEEEAPAYN DHLNFYFACE GLKQQTDPEK IKQIIGAIYR FLRKSQLSIS 120
121 DDLRAQIKAI KTNPEIPLSP HIFDPMQRHV EVTIRDNIYP TFLCSEMYIL YIQQMSAQQE 180
181 RCTSSGATGS GSAGSSGSGG SSLAGACALP PTTASGKQQL PQLVPPGAFI NLPVSSVSGP 240
241 PAGTCSASGS VYGPSTSASS SGSISATDTL PRSSTLPTLH EDSVLSLCDD FEKVQMQEGG 300
301 GSLGSGSVGA GARAPDYPIR LTRDLLIATQ KRRLEIRPPG AHGYVYNPST TNTSYVPNSR 360
361 VDSERASVSS GGRTDSDTMS ISSCSMDGRP YIQRRHSSTE SKAIRQSAMA NKETNTFQVI 420
421 PRTQRLHSNE HRPLKEEELV SLLIPKLEEV KRKRDLEERA RERNPGAALL TNERSSASDR 480
481 AFAEAIREKF ALDEDNDQDI LDQHVSRVWK DQTPHRSPGT MSPCPPIPSR RRTATHDSGM 540
541 VSDGAMSLSG HSMKHSKSMP DHSSCSRKLT NKWPSMNTDS GISMFSADTV TKYKDASTAS 600
601 KLEEAKRRLE DEPRRSRRYA QPPMQHLSQQ PLASFSSSSS GGSISLPHQP PPLPAKPPET 660
661 IVVFSFCEEP VPYRIKIPGT QPTLRQFKDY LPRRGHFRFF FKTHCEDPDS PVIQEEIVND 720
721 SDILPLFGDK AMGLVKPSD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
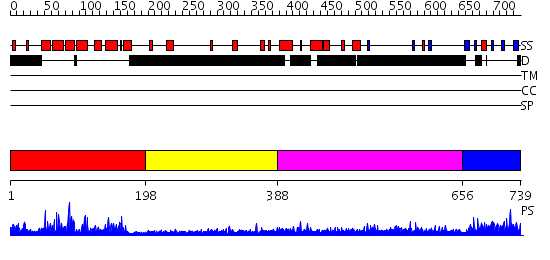
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..197] | 47.69897 | Regulator of G-protein signalling 4, RGS4 | |
| 2 | View Details | [198..387] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [388..655] | 1.217989 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [656..739] | 34.522879 | Crystal structure of axin dix domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)