













| General Information: |
|
| Name(s) found: |
Gycalpha99B-PA /
FBpp0084819
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 676 amino acids |
Gene Ontology: |
|
| Cellular Component: |
guanylate cyclase complex, soluble
[IDA][NAS]
cytoplasm [NAS] |
| Biological Process: |
cGMP biosynthetic process
[IEA]
intracellular signaling pathway [IEA] rhodopsin mediated phototransduction [IMP] positive phototaxis [IMP] |
| Molecular Function: |
guanylate cyclase activity
[IDA][NAS]
heme binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MACPFFRRAD SLTRQPSVIA EPGGHWALED EELSDDALTL THLQMAIQLL TAPSNEDLNT 60
61 AVTSLVAKYR QNWPNIHKLK LDPQTFKSCA NYDYLADIQE LLLKMDEASA SEILVLLGEE 120
121 LITCCCTGII ERAFRCLGTD LQEFLGSLDG VYDVLKLQEE DVTDTGFVCA GEGELIFTSE 180
181 RPVIAWLLLG SLKALTRMLY KVDVNIKIEP VEGDARRYRY LFSLVKDNSQ TMLMGRPTSV 240
241 SKTIPETVQR SNSSNASDLQ MNSSSFCKMF PWHFIMNEQL ELVQLGRGFS KLYKPYMADF 300
301 GCQATTYFDF KRPKGLTMKF RDIVRRTYTP FLIGLNNPPG AVDFPAIGLE IKGQMVHCPE 360
361 SNSLLFIGSP FLDGLDGLTC NGLFISDIPL HDATREVILV GEQARAQDGL RRRMDKIKNS 420
421 IEEANSAVTK ERKKNVSLLH LIFPAEIAEK LWLGSSIDAK TYPDVTILFS DIVGFTSICS 480
481 RATPFMVISM LEGLYKDFDE FCDFFDVYKV ETIGDAYCVA SGLHRASIYD AHKVAWMALK 540
541 MIDACSKHIT HDGEQIKMRI GLHTGTVLAG VVGRKMPRYC LFGHSVTIAN KFESGSEALK 600
601 INVSPTTKDW LTKHEGFEFE LQPRDPSFLP KEFPNPGGTE TCYFLESFRN PALDSELPLV 660
661 EHINVSMKTI SEGGDA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
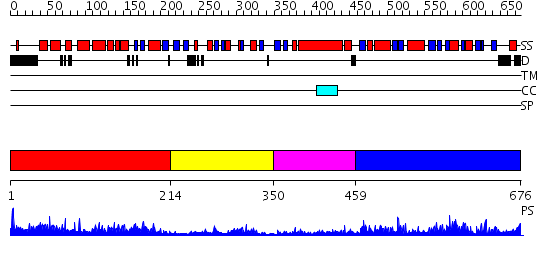
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..213] | 30.69897 | No description for 2o09A was found. | |
| 2 | View Details | [214..349] | 1.502996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [350..458] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [459..676] | 51.30103 | Crystal structure of a mutant form of the mycobacterial adenylyl cyclase Rv1625c |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)