













| General Information: |
|
| Name(s) found: |
CG1951-PA /
FBpp0084711
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 835 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein amino acid phosphorylation
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
protein kinase activity [IEA][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDMINKFYSS AVQTVSTLSG VLPGNNVTRE YEVLEQVCTA GVGLMWKVYN GHKKSTKQEV 60
61 SVFVFEKKSL ERWSKDDRET MLETLRRGVQ QLTKIRHPHV LTVQHPLEES RDSLAFATEP 120
121 VFASLANVVG DNVRSEKKLY DVEIRHGLLQ LFDGLQFLHQ DAKIVHRNIS AETIVINKNR 180
181 SWKLFGFDFC IANQPATDGT PHWPFREYTT SLHVLAQPSL EYTAPELALN SVNTPDSDLF 240
241 SLGVLIFTIY AGKPLKMFGS DYSSFRRYAN DLNQRKYPPM NAVPSELTES LKALLHPSAN 300
301 LRPKLHELKQ IAYFQDVGVK TLSYLDSLYQ WDNLQKSKFY KGLPQIIPTL PHRVNLHSIL 360
361 PYLVKEFVNS PMIPFVLPNV LLIAEMSSQR EYCDHILPHL KPIFKLTDPI QILLIFMQKM 420
421 DLLLKLTPAE EVKQSVLPLL YRSLECDMPQ IQDLCLAVLP TFSTLIDYNA MKNSVLPRIK 480
481 KLCLQSSNVS VKVNCLISIG KLLENLDKWL VLDEILPFLQ QIQSREPAII MGIIGIYKIA 540
541 MTNTKLGITK DVMAHKCIPY LVPLSVENGL TIAQFNTIIS LIKEMMGRVE QEQREKLQQL 600
601 STIQRDNKPK DASEILANEL ENSTLSPYGS INGNKINDDM FSGFTVGQPK AAIGAAIAPA 660
661 KAREQLKISH NTISAPPAAR PDILSSMMQS NLTGLGTITV AAAPPPTTTQ MPSNNGWHSA 720
721 NPLVMNHQLA SPQASNNNSF SLDDLDPFAG GRPSVGAPKA NNTNGANMYT VQQPTVNYIY 780
781 PTGYSLNSIQ QHQQQQLLGK PSQAPTLQPQ TQPLLPTDNQ NLNALSQQDI LNFLN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
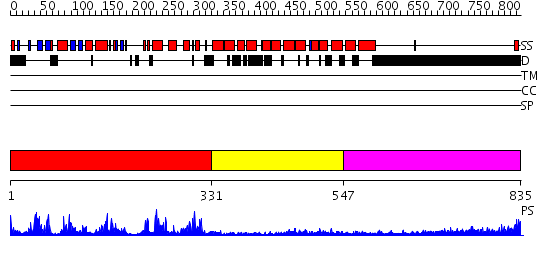
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..330] | 76.0 | No description for 2fh9A was found. | |
| 2 | View Details | [331..546] | 14.522879 | Regulatory apparatus of Calcium Dependent protein kinase from Arabidopsis thaliana | |
| 3 | View Details | [547..835] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)