













| General Information: |
|
| Name(s) found: |
FBpp0297117
[FlyBase]
FBpp0297116 [FlyBase] FBpp0297115 [FlyBase] FBpp0297114 [FlyBase] FBpp0297113 [FlyBase] mrt-PA / FBpp0084544 [FlyBase] |
| Description(s) found:
Found 52 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 775 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
olfactory learning
[IMP]
learning or memory [IMP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLMPRYLDSS TSSGSHGAAG ATNSSSSPPT TVAAHTKMRL LSKVEHNMRQ SQGHIQSNQS 60
61 NASGQDINHL LHTASTGNGV VDDVDGLLTP MRIKAHMSPP YSAGSSVKSD SLGYSPRPER 120
121 NLWSRAEMLE MLSIMQNLNA LEQLNDRNMK SEQVFRQIEE VMRSKGFVKK SSIQIWTKWK 180
181 FLKSTYNTTT RHGTGVPKVV PEEVYRVLCR MLSDTNHGGG NSNVNGSLSE CGNSMDSSKT 240
241 AGDDNKEDIL GVEHPIFGFR LGPIKPEPID IGYETTEKHD MVSEPDMESF DYEAGSSHAD 300
301 NGDQEDVPHL PFIVSVKHEP DVDLGMDTTN TPPPTAPASP SPPPVTVDDT EDDSVYSSTP 360
361 VAPPPLRVAS FAKGELRRPT HGILQIDRPP PTLTKMGQQQ IPNPGQMSRS VPLQSTSSIN 420
421 FVHPHSKLML PRRQNPNSSQ SGLRLPRDIS VQSTGMRMKT VPMRDTGYSL RPDRMTHDID 480
481 QSTSSPPQSP STMSSLLGRG PPPKYQIAGQ QAGASTSRQA HMMSSHQLTP VQHQQLQPRK 540
541 RRLGQSPVMG MPVPVKLQRS SNMYGSAPKD IRSHPAVGEE SKKKSSEEET KQRRKRQDEL 600
601 LAKELSQLAS AMRTAQKEML QDFFLQQKEI ARREHVFQMK QDSLVMRTLR RQTDALLRTA 660
661 NELVSGVSEA AKMEPDDKEG VIPETQMPFE PETELQTPES EEDNENGEGY AGDDVQETME 720
721 EPDYEDNEDD DYEHDEDEED EGDYDENMAN DSDLEMSNLA MSEGSNNAEI ISGGH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
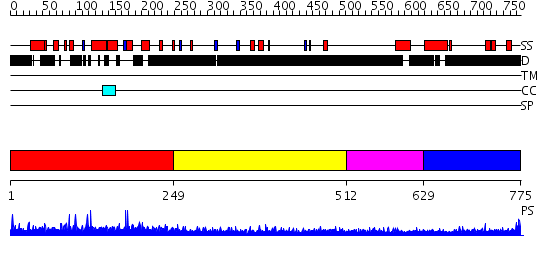
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..248] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [249..511] | 1.23299 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [512..628] | 5.237997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [629..775] | 1.190997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)