













| General Information: |
|
| Name(s) found: |
FBpp0310717
[FlyBase]
eIF4G2-PA / FBpp0088566 [FlyBase] |
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2072 amino acids |
Gene Ontology: |
|
| Cellular Component: |
eukaryotic translation initiation factor 4F complex
[ISS]
|
| Biological Process: |
RNA metabolic process
[IEA]
autophagic cell death [IEP] spermatogenesis [IMP] male meiosis [IMP] G1/S transition of mitotic cell cycle [IGI] spermatid differentiation [IMP] salivary gland cell autophagic cell death [IEP] translational initiation [ISS] meiotic G2/MI transition [IMP] |
| Molecular Function: |
translation initiation factor activity
[ISS]
RNA 7-methylguanosine cap binding [IDA] protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFPQQGGGGK GTRPQSTHQQ NHNHQAQHHQ ATHSHHHHLQ QQQEQQQSHA VFVPNHSGLH 60
61 HHQQQHLLQQ QQQQHQQQAA VAAAHLQQHI QQQQQQQVQA QAQAHHQPVI FYNTQPSYAA 120
121 HSAAVNNSAN GGGATLQLSA AASSNTLGHP LTGGHHGGAN GVAPGSNMPR ALSNPTLPPH 180
181 FQQSAGLIAP AGGNAGGPYI SATYIPINQT GPLMGMQPAV TFPTNGAAGA SFYAAAPPPP 240
241 QQKQLPPTAQ QQGPGGAPGA AQTPTLAYYQ LGPNAQNSAG NNVVSGANTH RGNMRGGQRS 300
301 TPTAQHTNYF TAYTMAPGNA QRPAPTNVAP MMNSRSGALN GAAAFHHLPH NFGIPSLALG 360
361 PPAGMTTPIL APAGMQAPPP QPPGGLHTFA PHLQSNSATT LPSVPSVTGV PAVASGFSTT 420
421 SLPTPSVTTA PQLGANSDKV CKRKYAIPII HPDTMENVMD GWKRSVFIKK DSNSTLTQHT 480
481 DLDTALDDNS TLCLYQAPTL MDSGSGSSLL SGAMVDQQMQ TVKPASSLKQ SVSLEKVPEA 540
541 VYCKDAVGQD VHPSPPPKST DKDQISVIVK DILAHSGNAE DHLSFWQYSD VDNVTETHNS 600
601 LPLVQSDLPD TTDTCQQPVL FQEQAKKQRP KLANKQSTAT PASIANDVAE AAASGCPGVS 660
661 ILSRGQSMVT PAGPSTPPPP SRVAQQKSTK SIVTTAVAAS VKPTTGSPNR KSSIQAATST 720
721 TTSSGASSST ASTPTHASSK LKQQPVNNLQ HGKQIPGKVS TPALPPTASR ISAAVTLLAG 780
781 TANITTTTNS SKKQRQAQKR ARELEKKKQK SSITSTTSTQ GQNNNGKGNG NGNAALEINT 840
841 NSNNAATVTT TSMSTLTTTT TATAPTTKQE TAAVDDKETA QPTTDTLSAT SNNTKVSSIL 900
901 RDKKTMLNPK EQLTSHNNNN NNNNEDGIEA KVESDPESDI KDDSMSNITG EKSEKHLANE 960
961 EIVAKFLQKD SPDTSVVHAE TQQLQFLNSS SSVCEEDEPV PVISEGVTKT EENAEAEIKF1020
1021 GDFHEDPRND TGFICSSTWS SSTISKAAED DSSQSSRSAD NVDCAPKVVE QISEEKSASS1080
1081 TKSSPVHKAT GSGPGSPKTK ELKSKSPSPI PVPQVPKNYR YSIETLRELA KRRDSRKPPM1140
1141 VPCQKGDCIS QLFVSRQQQP QQHHGQHIQQ YQHMSFNESL DYVPGKRGRG HGPNKKHHDQ1200
1201 HQQMGGSSGM GSGSGGGGGI SNSSQRHMDI IRVQLSLKEE IKLSECENAW QPGTLRRVSM1260
1261 AGGQDEDDDV EGVLKKVRGI LNKLTPDNFE VLLKEMSSIK MDNEAKMTNV MLLIFEKTIS1320
1321 EPNFAPTYAR FCKVLFHEIK AENKSLFTSS LITRIQHEFE SNVNDANAKS KKLQPTMERI1380
1381 NQCSDPAKKA ELRAEMEDLE YQFRRRAWGT VRFIGELFKL QSLTNDRVLN CVESLLEHGC1440
1441 EEKLEYMCKL LTTVGHLLES SLPEHYQLRD RIEKIFRRIQ DIINRSRGTS HRQQVHIKIS1500
1501 SRVRFMMQDV LDLRLRNWDQ PATHQSGQQG GRGQRHKQHD DSKDQSHSTG GGRGGGMNQH1560
1561 QQSSTHHQHQ KHNQHGHDGG NYFMQKMPNK QHHENQTLSI DPSKLRFSNS SATDDSIAKL1620
1621 GNSSHYQWRN VGNRPVSSTQ VNASNAPPPT SLLKRPGQNY KFLPYQQPSA VAGTANAPAG1680
1681 GNKSNVEVND AFDGKQCQGL ITKLVEEALN TREWQPEVLS IWRSHSGSQQ SKTLFHLLTD1740
1741 YLHKSTVKRQ HRQACGNIFA YLMDKDAVEK EIFEEAYSRF GDDFPDLLVD VPNGWGYVFE1800
1801 FLGPIIHSGH LDLKDVWQRR WLDDFFFAER FVYAFVSYFV QEFGAAYARK LWHNDYKLDR1860
1861 GQLFWSDVRK YREFVQSHSF YFLDNGPGQN QGHAKVKAPT TPRSPVEHVE RIRHLLAISC1920
1921 DMAIDYINTN VAINDKFVRQ LTRFLCCDFA LTVMTNTHNN IKSGGNSKPL QLNTESFRSC1980
1981 CTPLLRLCID AQEALEIACI DEAVDALQQH FMTEYEDDMA AGETICSTFS VLYESEVIPK2040
2041 ESFDKWYKLE MAHSRAYRRP FIDKLRGFIE EM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
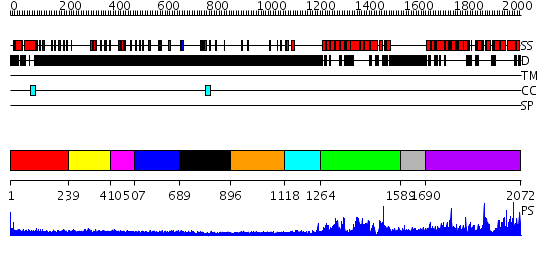
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..238] | 2.279996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [239..409] | 2.301988 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [410..506] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [507..688] | 2.296989 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [689..895] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [896..1117] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1118..1263] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1264..1588] | 111.0 | Eukaryotic initiation factor eIF4G | |
| 9 | View Details | [1589..1689] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1690..2072] | 121.0 | C-terminal portion of human eIF4GI |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. | ||||||
| 6 | No functions predicted. | ||||||
| 7 | No functions predicted. | ||||||
| 8 |
|
||||||
| 9 | No functions predicted. | ||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)