













| General Information: |
|
| Name(s) found: |
mbc-PA /
FBpp0083850
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1970 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ruffle
[TAS]
|
| Biological Process: |
dorsal closure
[TAS]
cell competition in a multicellular organism [IMP] cell morphogenesis [TAS] skeletal muscle tissue development [TAS] myoblast fusion [IMP][TAS] larval visceral muscle development [IMP] |
| Molecular Function: |
GTP binding
[IEA]
protein binding [IPI] GTPase binding [IEA] guanyl-nucleotide exchange factor activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVWSDCNAK QAEFGIAKCN FDQESKPHRL NLDVGDAVII LKETTHWYYG YRQKAKEIRG 60
61 IFPKSYIHLC EYNIVNGEYC IQRTDIVEEI TKVLLEWGSI AKDYFLTTNP SFPKIRRKMN 120
121 ELNNNRAALI SGNLPLDEVR KVKLLATNQI DTGNKLLGLD MVVRDESGDI LDTNAICTTE 180
181 LYEQHMHAVQ RIDKANRLSS ERGTTRTPNK YSHNILLHVN AFVCKFQEDS DLLFTLFDGE 240
241 THKPISENYV VKWSRTGIAR DTDQIDNNRV LFTDLSKSDL AIAKMYLVCY AIRIGSMEFK 300
301 DSAESKRTSM SIANSMLNAS SRKASQLSVS SSGSSSSNGE YIIRRPFGVA CKDLTPFINK 360
361 SDDFRGNIDL PFIMCEKETL DGTLRKLIAN KDIGKIDSKM AVTIEVLRGD IKQIKEEFPR 420
421 LMHTNVPVAR KMGFPEVILP GDVRNDLYLT ICSGEFARIA KTSEKNVEVS VCVANEQGYL 480
481 MPGVLSIGAG HQPIDEYKSV VYYHDDKPKW QETFKIHVPI EDFKQCHLRF VLKHRSSNEQ 540
541 KDRTEKPFGL AYVRLMQANG TTITQGQHIL AVYKIDHKKY DKTVANCYLE LPATVAELQG 600
601 AKPSIGGLTL LPKDQLSIGV NLCSTKLTQS VSLLGLLNWS AHKETLEQSL NALSTVPGEE 660
661 VVKFLQDILD ALFNILVEND HPEKYDQLVF MSIIHLIETV SDLKYQHFLS VLDVYINESF 720
721 SFTLAYTKLM DVLQKNISEA ISPKEKSADG NDLEESPEVR RLYKTTRYLH YVMKFVIRSR 780
781 VLYAEMNCNT DYMDFATRLQ ELLRMFIDMI GCPSNLLKSE GALLKNLHII ATDLMQVFEH 840
841 VRLSISIVEI LEKFPPRRLT QSKMGCIKDF VETKLFTLPK CRAILLPVFC KHIKDHLESK 900
901 EEIAECINIM NNILKLLFRS DVGSTHNDIR DIMIILFRTV MKAAHALDRD TGLVGKFFAI 960
961 MLGILQRMDA QHYEYFVKDL HQSGELKHFV IEILLVFEEL VSPHQKAVFP RDWMDMIMHQ1020
1021 NTVILGALKH LTVVITDYFL CPFEKQIWSN FFQCSIAFLV QSPLQLNDFN DNKRQIVFAR1080
1081 YRDIRKDTAM EIRKMWFQLG QHKPKFVPQL VEPILEMSMI PEKELRQETI PIFFDMMQCE1140
1141 YYSSRLEHES YGDTKFNNAH HKGNFSDFKT AMIEKLDILI GAGKGDAEYK HLFETIMLER1200
1201 CAAHNTLNVD GTAFVQMVTR LMDKLLEYRF IIQDESKENR MACTFSLLQF YSEVDLKEMY1260
1261 IRYVNKLCAL HMEFENYTEA AFTLKLHTEL LRWTDTELSH QLRSYRHNNC RTHRQLKEAL1320
1321 YFEIMEYFDK GKQWECAIDM CRVLARQYEE EIFDYLKLAE LLNRMALFYE KIIKELRHNS1380
1381 EYFRVCFYGR GFPRFLQNRV YIFRGKEYER HSDFCARMLV QHPQAELMQT LEAPGDDITN1440
1441 SDGQYIQVNK VEPIMGQAFN KFNDKIINNE IVKYFTANNV QKFQFSRPFR DSTNGGDRDD1500
1501 VRNLWLERTE LRISYPLPGI LRWFPVVETN TFKISPLERA VEIMKDTNRD IRQLVILHKS1560
1561 DETLHINPLS MKLNGIVDPA VMGGFAKYEE AFLTDDYLEQ NPDDKELVEE LKELIANQIP1620
1621 LLDLAIQLHR LRAPDSLKAL QEHLERCFAD MQQHVEQRYG RKSCDLKIER DSVVMRRPNS1680
1681 FLPSLFDGSN NRHSETSMGS SDSGLSKSTF LPRPQTNSIK NPFSGLSFNT RPSLGHSPSI1740
1741 KSNKSKDKTP SKRRTKDGKV KEREAHSLSS SQWYTPPLST ITSTPEKEIN TSIASLASTS1800
1801 NSSLSGPKTP DPHVLTEELT PKRPLRSEME KERRLSRPAS IATPTASIKN FPDTRSLSES1860
1861 SNRNSVETTD STSEEDIRPP PLPAKARDST DFTSLSQNMD WTPNGYAMLS TISNTSSMST1920
1921 TSTLTKTSIT NTTYEYLETT NFSLVGAIDG NKPRPPTPPP KPSRHSKHIP |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
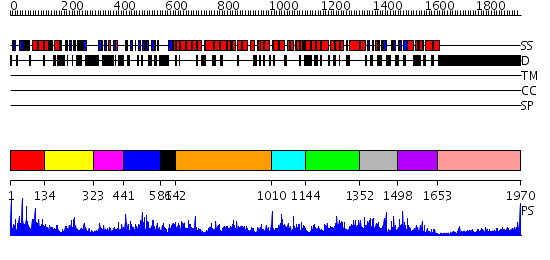
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | 5.69897 | No description for 2dl3A was found. | |
| 2 | View Details | [134..322] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [323..440] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [441..580] | 1.04 | No description for 2dmgA was found. | |
| 5 | View Details | [581..641] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [642..1009] | 3.30103 | Crystal structure of the exportin Cse1p complexed with its cargo ( Kap60p) and RanGTP | |
| 7 | View Details | [1010..1143] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1144..1351] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1352..1497] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1498..1652] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1653..1970] | 1.58 | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)