













| General Information: |
|
| Name(s) found: |
cdi-PA /
FBpp0083130
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1213 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
embryonic development via the syncytial blastoderm
[IMP]
protein amino acid phosphorylation [IEA][ISS][NAS] |
| Molecular Function: |
ATP binding
[IEA]
protein tyrosine kinase activity [IEA] protein kinase activity [NAS] protein serine/threonine kinase activity [ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLNQHSGPG PPSEICGLSV DQVDSSAASP FIWPSTAGMC TTATTVTTST NGDATAEAPV 60
61 KHQPLHNGGW IGNGLPPAPV TRTISSDRLV TGSSCRALRT AVSALYSVDD FVKEKIGSGF 120
121 FSEVYKVTHR TTGQVMVLKM NQLRANRPNM LREVQLLNKL SHANILSFMG VCVQEGQLHA 180
181 LTEYINGGSL EQLLANKEVV LSATQKIRLA LGIARGMSYV HDAGIFHRDL TSKNVLIRNL 240
241 ANDQYEAVVG DFGLAAKIPV KSRKSRLETV GSPYWVSPEC LKGQWYDQTS DVFSFGIIQC 300
301 EIIARIEADP DMMPRTASFG LDYLAFVELC PMDTPPVFLR LAFYCCLYDA KSRPTFHDAT 360
361 KKLTLLLEKY EHESSAGASP LSSLELINCS PGGSDSENVT LTPRASVPTV ATICTAPAGT 420
421 LTPRHLQEVD EDGFRFPSKK SASAALPGTP NGSAGSSGFD DHILKSRVRR ATQKLEAEIL 480
481 LHRRSLSENI IHFPMHTSPS DKARCHQMQR QRSSTHSPTP PRQLSSVQLP ATSIPDPPPL 540
541 PKPVPAVLTL RKVGDPQYKP TSEKDHNGSK PNPFEALAQL RGVKKILDAN PKTYAAGVGD 600
601 LFSSCFEMLA PFFKELAAMQ SKAGAQGGGT STEETSTGNK ASAATPSEPK SLPVSPHMTR 660
661 KYSAIELRLP QRISNAAPPA VTSAPPPSIA LPTDEDDCDD CNSEGEDPDG GGNCADAESS 720
721 SDDSAVRPST VPTARKYRAN SLYAHPLFRG SNGTSMKSTS TGEFESPTLL KASASTITLT 780
781 ASVGATNSCE DLTESARKSK EQASSGATAA VQALPTPMSG CSTRLVETPV SVSSCTDLEL 840
841 DELKNTCDLN ELKNSCEQLP LKTNLTRRDS IESGFFSCFN EESDAAITNN SFGRQLNGIG 900
901 SLGFGCGGGG NSSNCCYEKL KTQLMLDACV SADSEINDKL ASLCRCCYYA TSTSNAAVTA 960
961 AAAAAQQQQQ SHLLNDTSST SSSVLLIDND PAVIGGNTSS STALDSMDDL DADIVASRRT1020
1021 HQRRYTCHHA LLSGNGSESQ MAATASLINR LALDSEIHTL LQRSPFATNQ LLYCKNRTSS1080
1081 IYTDSSDDIS SLAGSDSLLW DDRSFTALPS TRSAQIAKIV EYFERKRQDS SPPSVHHRNG1140
1141 NGTVAAAVAA SALGSNYLSY ESRRYSDFKR HLNADGLCFD LDKKPAQQRM MVCEGAVKSK1200
1201 LPIFDKKKQQ QGG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
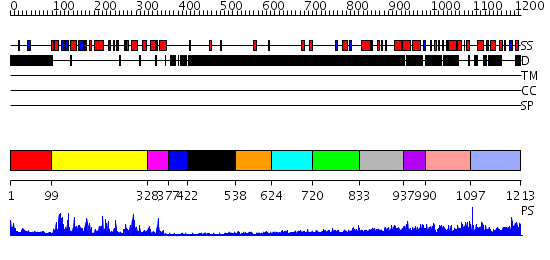
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | 87.045757 | Abl tyrosine kinase, SH3 domain; Abl tyrosine kinase; Abelsone tyrosine kinase (abl) | |
| 2 | View Details | [99..327] | 30.69897 | No description for 1wakA was found. | |
| 3 | View Details | [328..376] | 3.1 | No description for 2ivsA was found. | |
| 4 | View Details | [377..421] | 75.154902 | Catalytic and ubiqutin-associated domains of MARK2/PAR-1: Wild type | |
| 5 | View Details | [422..537] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [538..623] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [624..719] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [720..832] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [833..936] | 10.39794 | Solution structure of the F-actin binding domain of Bcr-Abl/c-Abl | |
| 10 | View Details | [937..989] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [990..1096] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1097..1213] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. | |||||||||
| 7 | No functions predicted. | |||||||||
| 8 | No functions predicted. | |||||||||
| 9 | No functions predicted. | |||||||||
| 10 | No functions predicted. | |||||||||
| 11 | No functions predicted. | |||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)