













| General Information: |
|
| Name(s) found: |
CG7156-PA /
FBpp0083026
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 681 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cell communication
[IEA]
protein amino acid phosphorylation [IEA] |
| Molecular Function: |
ATP binding
[IEA]
protein binding [IEA] phosphoinositide binding [IEA] protein kinase activity [IEA][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTLNLGGWIH SFTVTDTQEH KGGYTIYKIT SIVFPRSVPQ ALTCLVVWKR FHDVKRLHRE 60
61 LSRRHKSLQL PGKLPVPTDS TYFKRFDAAV IQRRKEYILQ LLDFAAQHPA LYKCSTFTQF 120
121 FQETPSPNGS PLKKLYVERS LRLQTEDNGC GSGGSETGTC AGAIADICDK LDLPYDPHDA 180
181 GGITPLDPRC DKEQTNNESQ AKETEIELET KQEPKTDVAL TKRDYLRLLT PMASIESDDS 240
241 DYIYEAALEF SHAVQAEVNL EYAEAHERYK HGVDLLLNGA KQDSSEERRF IAKAKIAKYL 300
301 ARAEEIHANF LANDCPTKKL QFQLSLDTTD GGSVSHLECP WNQLARYKVL QILQDRVMQV 360
361 QCVTKSQQPA AIMKGVEKPA SHSLTQSIFL PQQVPYMVQL LAYFQSEQKI FLLLRQAEGG 420
421 MLYDYLQSYT PTSSKSVNYG ELFPSDQEPE EKEAEEPPLT SDSDVSDLVR SSQQLLKSVT 480
481 NTLSNLQAGV VTPKRNKKVD QNRIRSKPIP EQCLRQWACE LAIAIHSLHG KGVILRDLHM 540
541 GNILLGAKGQ LLLTYFYQNE GLTSDDNYVH KALNLEAVAN HFVAPERPLT FRSDWWSYGV 600
601 VLYQLLLGVP YKVTHPGQLD LYGCVQYPSE SLEISDHAKD LLEQLLQLSP ELRLDYDQLQ 660
661 AHPFFKGIDW QAVLEQGTES M |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
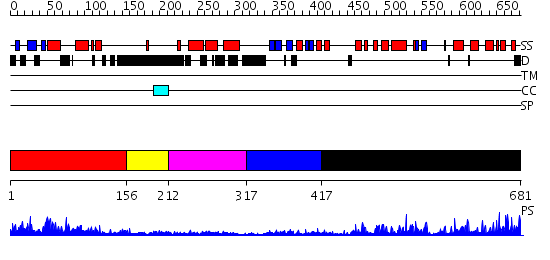
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..155] | 23.0 | crystal structure of CISK-PX domain | |
| 2 | View Details | [156..211] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [212..316] | 4.07 | No description for 2v6xA was found. | |
| 4 | View Details | [317..416] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [417..681] | 66.30103 | Pkb kinase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)