













| General Information: |
|
| Name(s) found: |
Gyc-89Da-PC /
FBpp0110263
[FlyBase]
Gyc-89Da-PB / FBpp0110262 [FlyBase] |
| Description(s) found:
Found 39 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 667 amino acids |
Gene Ontology: |
|
| Cellular Component: |
guanylate cyclase complex, soluble
[ISS]
|
| Biological Process: |
cGMP biosynthetic process
[IEA]
intracellular signaling pathway [IEA] |
| Molecular Function: |
guanylate cyclase activity
[ISS][NAS]
heme binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYGMLYESVQ HYVQEEYGVD IWRKVCHIID CKHNSFKTHQ IYPDKLMPDI AEALSACTGE 60
61 SFDFCMNFFG RCFVRFFSNF GYDKMIRSTG RYFCDFLQSI DNIHLIMRFT YPKMKSPSMQ 120
121 LTNMDDNGAV ILYRSSRTGM SKYLIGQMTE VAREFYGLEI KAYVIESQND ISGGTAGPIK 180
181 LTDGPLTVIV KYRLDFDNRE YMAKRVNTEA HPSQLKMPTV KLDVFLDLFP FTFVLNHDMK 240
241 ITHAGEKIVE TWIMHNPGAN PKSFIGTHVM DLFQCRRPKD TTIDWDTLIQ MRAVLFEFEL 300
301 IRTGHNRAAY DAVLNMDFEN YDEMDLNEAQ TMALAKAQEF SESHPVDDDE SAREDEIDPA 360
361 TGERRSSQGL RSILLKGQMF YIKDVDSLIF LCSPLIENLD ELHGIGLYLN DLNPHGLSRE 420
421 LVMAGWQHCS KLEIMFEKEE QRSDELEKSL ELADSWKRQG DELLYSMIPR PIAERMRLSE 480
481 EQVCQSFEEV SVIFLEVMNV YDEGLNSIQG AMQTVNTLNK VFSALDEEII SPFVYKVETV 540
541 GMVYMAVSGA PDVNPLHAEH ACDLALRVMK KFKAHDMGDV AIRVGINSGP VVAGVVGQKV 600
601 PRYCLFGDTV NTASRMESSS DPWKIQLSKY TGDKVRQVGY KVESRGTVQV KGKGDMETYW 660
661 LLEGPEG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
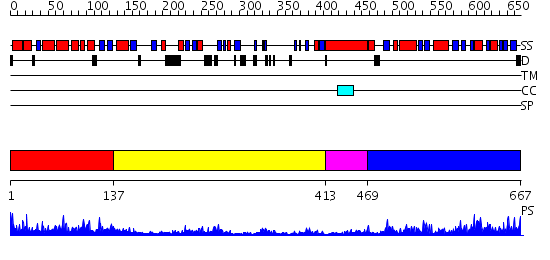
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..136] | 42.30103 | No description for 2o09A was found. | |
| 2 | View Details | [137..412] | 4.69897 | No description for 2oh4A was found. | |
| 3 | View Details | [413..468] | 4.0 | The structure of p38 alpha in complex with a dihydroquinolinone | |
| 4 | View Details | [469..667] | 50.69897 | Crystal structure of a mutant form of the mycobacterial adenylyl cyclase Rv1625c |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)