













| General Information: |
|
| Name(s) found: |
gi|7300134
[NCBI NR]
gi|24647409 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 925 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
bristle morphogenesis
[IMP]
|
| Molecular Function: |
nucleic acid binding
[IEA]
zinc ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAAGSNVRC INYYDLCRIC TDSSDHKTNI YSPEGRAKNL SQKILECLSL QVIDEKDRLP 60
61 KVVCAQCVQL VEQIHGLRAT CRNSQTMLNN CLNIRPAPSS DKLYIRDAVD ETGKSLSVAQ 120
121 SAPPVPNGAQ QSGNLLNSII QAVNMQPQQQ YTITMDNGVQ QQQQQHQAMP EVQIKSERQT 180
181 ALEEFIRLKP DIKITPLGKK EPISPAKPQQ QHSLQSQTVT PTINPTQLQL QPQLGDQLQP 240
241 LWQQIQQLQL QQQLQQLTNQ LQATTQYSVA QDDTSSSGDS KKPKLNFVLS SPTAPQFTTS 300
301 MPQFGGSQPQ IQLQLMPGSG GVTSAGTVPA QTQLNAAALL SSLAGPQSTT NLLSPNKCFL 360
361 PITIRDENSD QQIVAHIDTK NLVLPTTYQV QMKLQPQLAT ADGQPIMQLT PTSIPATLQL 420
421 TPQTLGNPGS AFQGTTVTQN QFLAPQQPLT QLPNTAQVTS QQIIRSPHTS TTNSQLVIRN 480
481 VTNIPPTPTT PTSSKKAPTF KTPSPKQKPM PSPKSKTTVG PSSGGSNGTS TNEFKRLITQ 540
541 TKPQPQVKQQ QTAGLSASGA PTATSANQAA MQRLSSNTTI TKVPKQPASL PAPAPTSNPA 600
601 KLPMLNKQNI TISRISMQTA PKAQSKPSTT SPTPASQPIP VSVPALSQAQ PPPLAAQHKK 660
661 IVRKPPENQD TSGQKVKAAR PPQQILPSPQ QEGQNATHSG SEAPTTSGLI CPTCKREFKK 720
721 KEHLTQHVKL HAGLRPFKCS EEGCDKTFSR KEHLSRHLVS HSGQKMYTCE VCKKPFSRKD 780
781 NLNKHRRIHT QTSTETLYCC DVCNKNFATK LHYEKHREMH KKIRPESAAP GATASPAAAP 840
841 ALTHVRSNGQ VPNKAVFEIK QERSAQQTQT QQPPAQVMHV VTTQDLAGNT ITITQAPDSN 900
901 MPGSLANYVQ LGFSQFQNPS ASVAK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
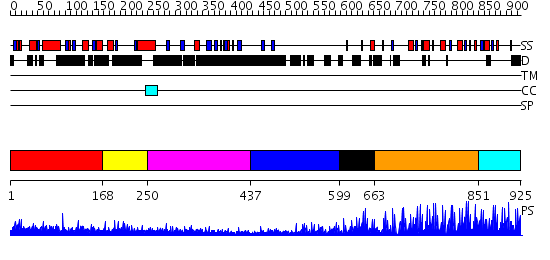
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | 2.59 | Crystal structure of the zinc finger associated domain of the Drosophila transcription factor Grauzone | |
| 2 | View Details | [168..249] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [250..436] | 1.129996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [437..598] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [599..662] | 1.569995 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [663..850] | 55.154902 | No description for 2i13A was found. | |
| 7 | View Details | [851..925] | 1.580997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. | |||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)