













| General Information: |
|
| Name(s) found: |
gi|24645763
[NCBI NR]
gi|23170940 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 614 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial large ribosomal subunit
[ISS]
|
| Biological Process: |
translation
[ISS]
|
| Molecular Function: |
structural constituent of ribosome
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRLTNTLCAQ HIGWHFKKHW LVQGRRVPKE TGAAAELLKL GVLVKNPEDL LNAKVEKKLV 60
61 DIVGAREKPV PEDSRHPDWH PTVCNTYSDN SVLIGGLPQA QVLTNSIVIQ TFPKHIEDAI 120
121 ASQQLPNSVD KSVRHAILAS HVLDAEQVKL PKVRLPERPA FNLPRSYGIS HERVNRLLVN 180
181 KLLHESEKLA GRSVSVRRKL IDNASFKTFL SKDGDLLGFS INAEKVVFAN RAIEGVKGKF 240
241 EGDLPDLYPM KSTISIPKYH IYQAKNLYPL RSDITCSHPH TIFTVFNKHQ VKNSHGSEVT 300
301 TSQLQARTLV KAFVVAAARA KQLHGDSVGA LPKPIVVQSV QTDGRTFHFG VLQLNTLDLG 360
361 ANSTAKNYWF HRQNYNLQKQ STSSNNSGTS SETSETQSVD DQQGTPHSAI RSSSASLSRA 420
421 KNSWRRMKEL AAEKEKENEA KRPPWRAVSV STLTKPDKSA LLRAKLLDAS RRLRAGKANV 480
481 ALQTDFVPTK LMKEVSFGVQ KDLILLKDVG ILTDGQYSKK KDGYEKYVLT YSMSQMTDSV 540
541 PTVSRQTQTL LPKAYNKDLV NSYLPKSDED DSDGISDVEK RLGDQIAKIR RLNFDESNLS 600
601 RGKNRLVRPA AVHR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
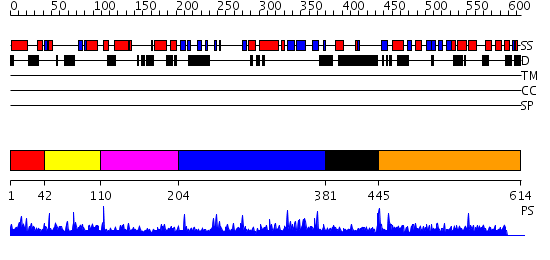
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..41] | 1.010989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [42..109] | 1.02499 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [110..203] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [204..380] | 2.031975 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [381..444] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [445..614] | 1.00497 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)