













| General Information: |
|
| Name(s) found: |
trbd-PA /
FBpp0081569
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 778 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: |
positive regulation of Wnt receptor signaling pathway
[IGI]
|
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCDTKDDAQK WKCETCTYEN YPSSLKCTMC QASKPLLNED IFRLSPAQES CTVAEEAAAV 60
61 EVAVMSPTPS STCYSLQPQS QARQSNVADS EKWPCKVCTY LNWPRSLRCV QCCTKRGGEA 120
121 IERGKKDMDN EADGDRAGEA LQALRISGSE ENLANKPVQL IGATASHRLS LSRGIDDATH 180
181 LNNLANASHN QSQSQHRQPV LQQQMQLQLQ PQQQRESSSS AAVPPQQQKQ CYVSKWACNS 240
241 CTYENWPRSI KCSMCGKTRE REISGSQNDL HASSSLNSQE ENQQQLQQPN VDTVSVNNSF 300
301 NKKHIYQLGS SETINNCDTL QERQERRQRQ IRRQVDWQWL NACLGVVENN YSAVEAYLSC 360
361 GGNPARSLTS TEIAALNRNS AFDVGHTLIH LAIRFHREEM LPMLLDQISG SGPGIKRVPS 420
421 YVAPDLAADI RRHFANTLRL RKSGLPCHYV QKHATFALPA EIEELPIPIQ EQLYDELLDR 480
481 DAQKQLETPP PALNWSLEIT ARLSSRMFVL WNRSAGDCLL DSAMQATWGV FDRDNILRRA 540
541 LADTLHQCGH VFFTRWKEYE MLQASMLHFT LEDSQFEEDW STLLSLAGQP GSSLEQLHIF 600
601 ALAHILRRPI IVYGVKYVKS FRGEDIGYAR FEGVYLPLFW DQNFCTKSPI ALGYTRGHFS 660
661 ALVPMEPFTR IDGRRDDVED VTYLPLMDCE LKLLPIHFLT QSEVGNEESM MRQWLDVCVT 720
721 DGGLLVAQQK LSKRPLLVAQ MLEEWLNHYR RIAQVITAPF IRRPQITHYS SDGDSDEE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
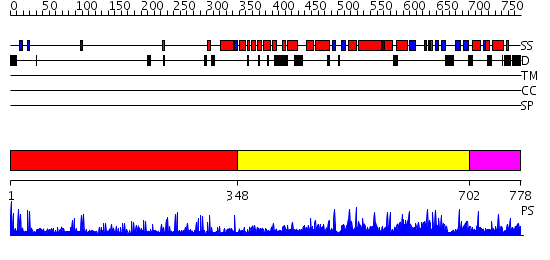
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..347] | 8.0 | Low density lipoprotein (LDL) receptor YWTD domain; Low density lipoprotein (LDL) receptor, different EGF domains; Ligand-binding domain of low-density lipoprotein receptor | |
| 2 | View Details | [348..701] | 1.1 | ESCRT-II | |
| 3 | View Details | [702..778] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)