













| General Information: |
|
| Name(s) found: |
hb-PA /
FBpp0081432
[FlyBase]
hb-PB / FBpp0081431 [FlyBase] |
| Description(s) found:
Found 40 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 758 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS]
intracellular [IEA] |
| Biological Process: |
torso signaling pathway
[IGI]
regulation of cell fate specification [TAS] posterior head segmentation [TAS] terminal region determination [IGI] anterior/posterior axis specification, embryo [NAS] regulation of development, heterochronic [TAS] anterior region determination [NAS] positive regulation of transcription [IDA] neuroblast fate determination [IEP][IMP] trunk segmentation [TAS] salivary gland development [TAS] zygotic determination of anterior/posterior axis, embryo [NAS][TAS] regulation of transcription [NAS] open tracheal system development [TAS] ganglion mother cell fate determination [TAS] morphogenesis of a branching structure [TAS] ventral cord development [NAS] epithelial cell migration, open tracheal system [TAS] central nervous system development [TAS] |
| Molecular Function: |
DNA binding
[IDA]
SUMO binding [IPI] zinc ion binding [IEA] specific RNA polymerase II transcription factor activity [NAS] transcription activator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQNWETTATT NYEQHNAWYN SMFAANIKQE PGHHLDGNSV ASSPRQSPIP STNHLEQFLK 60
61 QQQQQLQQQP MDTLCAMTPS PSQNDQNSLQ HYDANLQQQL LQQQQYQQHF QAAQQQHHHH 120
121 HHLMGGFNPL TPPGLPNPMQ HFYGGNLRPS PQPTPTSAST IAPVAVATGS SEKLQALTPP 180
181 MDVTPPKSPA KSSQSNIEPE KEHDQMSNSS EDMKYMAESE DDDTNIRMPI YNSHGKMKNY 240
241 KCKTCGVVAI TKVDFWAHTR THMKPDKILQ CPKCPFVTEF KHHLEYHIRK HKNQKPFQCD 300
301 KCSYTCVNKS MLNSHRKSHS SVYQYRCADC DYATKYCHSF KLHLRKYGHK PGMVLDEDGT 360
361 PNPSLVIDVY GTRRGPKSKN GGPIASGGSG SGSRKSNVAA VAPQQQQSQP AQPVATSQLS 420
421 AALQGFPLVQ GNSAPPAASP VLPLPASPAK SVASVEQTPS LPSPANLLPP LASLLQQNRN 480
481 MAFFPYWNLN LQMLAAQQQA AVLAQLSPRM REQLQQQNQQ QSDNEEEEQD DEYERKSVDS 540
541 AMDLSQGTPV KEDEQQQQPQ QPLAMNLKVE EEATPLMSSS NASRRKGRVL KLDTLLQLRS 600
601 EAMTSPEQLK VPSTPMPTAS SPIAGRKPMP EEHCSGTSSA DESMETAHVP QANTSASSTA 660
661 SSSGNSSNAS SNSNGNSSSN SSSNGTTSAV AAPPSGTPAA AGAIYECKYC DIFFKDAVLY 720
721 TIHMGYHSCD DVFKCNMCGE KCDGPVGLFV HMARNAHS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
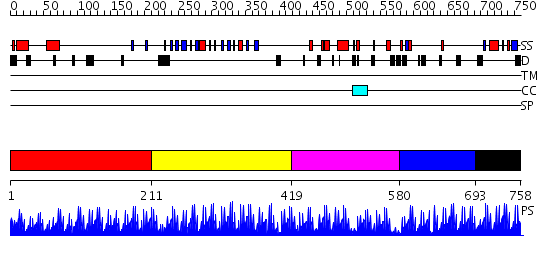
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..210] | 1.46297 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [211..418] | 58.0 | No description for 2i13A was found. | |
| 3 | View Details | [419..579] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [580..692] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [693..758] | 8.522879 | ADR1 DNA-BINDING DOMAIN FROM SACCHAROMYCES CEREVISIAE, NMR, 25 STRUCTURES |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)