













| General Information: |
|
| Name(s) found: |
FBpp0308676
[FlyBase]
DNApol-iota-PA / FBpp0081294 [FlyBase] DNApol-iota-PB / FBpp0081293 [FlyBase] |
| Description(s) found:
Found 42 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 737 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
translesion synthesis
[IDA][ISS][NAS]
|
| Molecular Function: |
DNA-directed DNA polymerase activity
[IDA][IMP][NAS]
damaged DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDFASVLGKS EAHQRTIIHL DMDYFYAQVE EIRDPTLRSK ALGIQQKNIV VTCNYVARAK 60
61 GVTKLMLIAE AQRICPDLVL VNGEDLAPYR QMSQRIFDLL LNYTPLVEKL GFDENFMDVT 120
121 ALVELRQAHV AEALLRPPVG HTYPADGTPL SNCDCGCAQR LAIGTRIAQE IREELKLRLG 180
181 ITCCAGIAYN KLLAKLVGSS HEPNQQTVLV STYAEQFMRE LGDLKRVTGI GQKTQCLLLE 240
241 AGMSSVEQLQ QCDMDVMRKK FGFETATRLR DLAFGRDTSL VRPSGKPKTI GMEDACKPIS 300
301 VRTDVEERFR MLLKRLVEQV AEDGRVPIAI KVVLRKFDSQ KKSSHRETKQ ANILPSLFKT 360
361 SMCPGETGVS KVQLADGAQD KLLKIVMRLF ERIVDMSKPF NITLLGLAFS KFQERKVGSS 420
421 SIANFLIKKA DLEVQSITSL TNTSLTSPTA ESPTSDECAF RSSPTTFKPS DQFYRRRATT 480
481 ASPVPMLLDN GSESAATNSD FSDFSETEVE PSPKKSRIGR LLVSKRSRLA ADVGDSAAEV 540
541 ASPSKLRVCD LRLNSRDSEK DFPMSTTPST STSAPAPRFR TVQPPNTLLQ RIDGSLRFVT 600
601 TRTASRLSSN ASSTASSPLP SPMDDSIAMS APSTTTLPFP SPTTTAVVTS SSSTATCDAL 660
661 TNIVCPAGVD AEVFKELPVE LQTELIASWR SSLVAAVEQT NGTGAATSAA IASGAPATAT 720
721 TASGQKNTLY RYFLRNK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
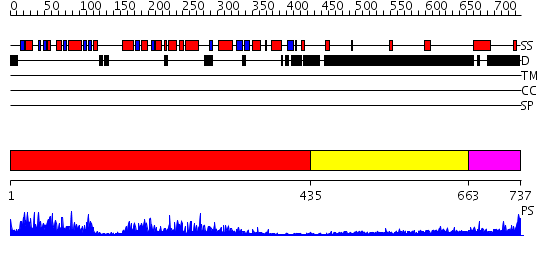
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..434] | 97.0 | No description for 2dpiA was found. | |
| 2 | View Details | [435..662] | 2.248996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [663..737] | 2.219995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)