













| General Information: |
|
| Name(s) found: |
Hpr1-PA /
FBpp0078316
[FlyBase]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 701 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleoplasm
[ISS]
|
| Biological Process: |
mRNA export from nucleus
[NAS]
signal transduction [IEA] |
| Molecular Function: |
protein binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTAVEVANT APEKLANYFA LQSAFEKALE LAITDGKVEL LVKEYNRFPA NTEHDKRLPM 60
61 DHAFRVLLMK RLDEDVSRIG ELVRLSVEAT RAEIVSNTIP VVLLIDTFDV VTLDKCQKIF 120
121 QFVEDMVEVW KEEIFFSSCK NNILRMCNDL LRRLSRTQNT VFCGRIQLFL SKFFPFSERS 180
181 GLNIVSEFNL DNFTEYGLDS KDHDESDNKE LEDTAEDIPL KIDYDLYCKF WSLQDFFRNP 240
241 NQCYNKPQWK MFQMHADNIL QSFSSFKLED VRQSSNENAS GTDQAMDVDE EAVDTTAVSS 300
301 VIKANHFFAK FLTNPKLLAL QLSDANFRRA VLVQFLILFQ YLQVSVKFKS DTQTLTADQA 360
361 DFIKETESRV YKLLEETPPY GKRFSRTVYH MLAREEMWNN WKNEGCKEFK KPEEPTLSEE 420
421 DSKPTPNKRP RRPLGDALRD ASRSGKFYLG NDNLTRLWNY SPDNLQACKS EQRNFLPLLE 480
481 TYLETPHEKV DPAFEWRALR LLARQTPHFF TSLSQPSSKI SDYLEQVRKR LIRDKEPKPA 540
541 ALLSNNSSEN NFGLSVNNVH TEGEQDATVA LQDGEPEQDL EVEDNEPVVE EVDEVPAHEK 600
601 PLMVTKEHIE EVAPLIGEPW MKVGKKIGFT NDELLFFQME HPTANVACVK MLTNWISDDD 660
661 DATLENWAYM LEGLEMNKAA EAVKAIIERE KSGSTAAFCL R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
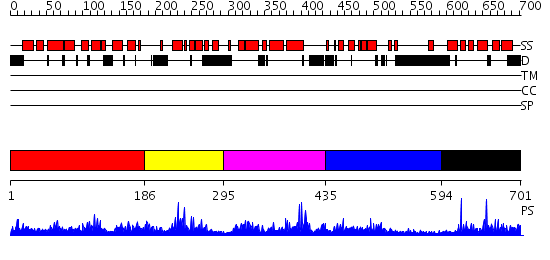
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..185] | 2.035958 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [186..294] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [295..434] | 1.034974 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [435..593] | 1.106993 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [594..701] | 23.522879 | Solution structure of the death domain of nuclear matrix protein p84 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)