













| General Information: |
|
| Name(s) found: |
hd-PA /
FBpp0078445
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 568 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][NAS]
|
| Biological Process: |
cell proliferation
[IMP]
S phase of mitotic cell cycle [NAS] DNA amplification [IMP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTEASVASK WTRPDDFIKL QRLKQKKNKL AARVSNNNNR RPRHQVDETD KSKLLEAKLA 60
61 QKRKNPFAKS TADAKKLRVD PQLADLEPVV TASSCFVRET PTRPAPAKFV LQKYDPQAFA 120
121 KLFQHPQINA EDEDDAELAA RQKHTAHLPV DWSLKTRARF FCPTELPAIQ LKTSQLASGL 180
181 TSFVRCMDPQ RTESTLDISD ATRFNQCNYY WQHPHLPWLT LYPRTAKENV GVVVGERERK 240
241 ALAEEWDFSF RGLFQLLRAR QCPYFYLCAN TFTVLFRAAG VGGRPESHAL VTPSTRGMRQ 300
301 ALRQEGIEFS MPLKSDNSGN AHDNSFNEES TTTSLGPEAG EDAPPPAQED EDDDEDWLES 360
361 LGVDERELRR IQSSHARKQQ AAEMREDFSD NSLLLVDGVE CQGFFSYLLN AKSAISTVGR 420
421 LAGVPPTLLS PVAFPKATMQ HLVPRSKKVR LDGVDYFSID IKGLILPTFL PSVAELLSET 480
481 RQMFSSTLAS SINTLAFSKA TQKLLETPET PQSDAEGEDA AGQVFGEQNL SECGLLPAVV 540
541 GSICRTGQHA VGLLERVCYQ RDEGYAWS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
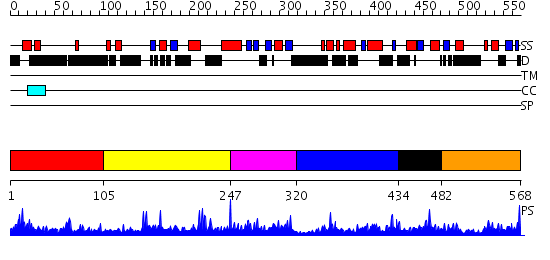
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..104] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [105..246] | 1.021959 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [247..319] | 1.021979 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [320..433] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [434..481] | 2.018992 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [482..568] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)